Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
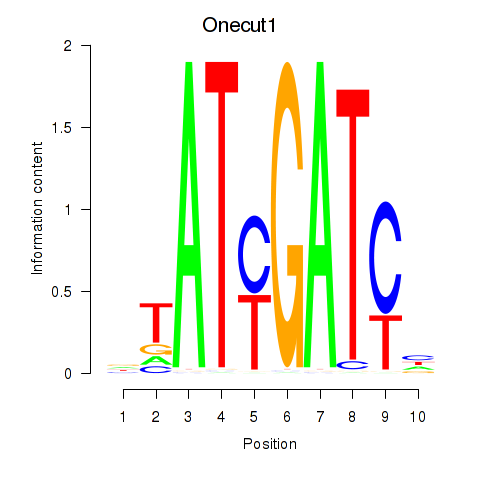
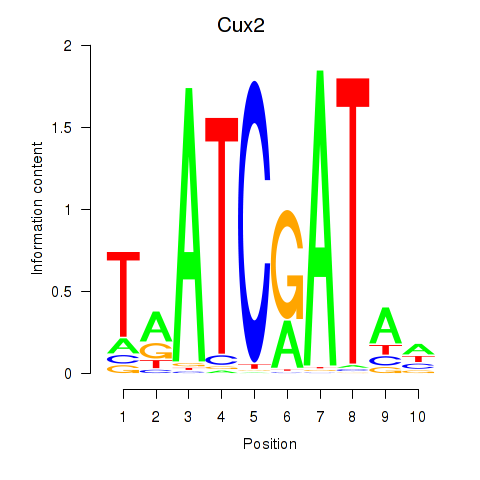
Results for Onecut1_Cux2
Z-value: 0.08
Transcription factors associated with Onecut1_Cux2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Onecut1
|
ENSRNOG00000008095 | one cut homeobox 1 |
|
Cux2
|
ENSRNOG00000001259 | cut-like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cux2 | rn6_v1_chr12_+_40018937_40018937 | -0.71 | 1.8e-01 | Click! |
| Onecut1 | rn6_v1_chr8_+_81766041_81766041 | -0.65 | 2.3e-01 | Click! |
Activity profile of Onecut1_Cux2 motif
Sorted Z-values of Onecut1_Cux2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_108415093 | 0.02 |
ENSRNOT00000031650
|
Syndig1l
|
synapse differentiation inducing 1-like |
| chr6_+_135856218 | 0.02 |
ENSRNOT00000066715
|
RGD1560608
|
similar to novel protein |
| chr15_+_50891127 | 0.02 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chr4_-_44136815 | 0.02 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr6_+_8669722 | 0.02 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr5_+_148051126 | 0.02 |
ENSRNOT00000074415
|
Ptp4a2
|
protein tyrosine phosphatase type IVA, member 2 |
| chrX_-_138148967 | 0.02 |
ENSRNOT00000033968
|
Frmd7
|
FERM domain containing 7 |
| chr2_-_96032722 | 0.01 |
ENSRNOT00000015746
|
LOC100360846
|
proteasome subunit beta type 6-like |
| chr17_-_9234766 | 0.01 |
ENSRNOT00000015893
|
AABR07026997.1
|
|
| chr2_-_98610368 | 0.01 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr14_-_34218961 | 0.01 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr4_-_50312608 | 0.01 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr3_-_47025128 | 0.01 |
ENSRNOT00000011682
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr2_+_114413410 | 0.01 |
ENSRNOT00000015866
|
Slc2a2
|
solute carrier family 2 member 2 |
| chrX_+_131381134 | 0.01 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr4_-_170912629 | 0.01 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr17_+_9736577 | 0.01 |
ENSRNOT00000066586
|
F12
|
coagulation factor XII |
| chr18_-_59819113 | 0.01 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr18_-_52047816 | 0.01 |
ENSRNOT00000018368
ENSRNOT00000077491 |
LOC100174910
|
glutaredoxin-like protein |
| chr19_+_60017746 | 0.01 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr19_+_23389375 | 0.01 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr3_-_72171078 | 0.01 |
ENSRNOT00000009817
|
Serping1
|
serpin family G member 1 |
| chr3_+_37545238 | 0.01 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr5_-_99033107 | 0.01 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr17_-_42267079 | 0.01 |
ENSRNOT00000059564
|
LOC100911664
|
uncharacterized LOC100911664 |
| chr11_+_13499164 | 0.01 |
ENSRNOT00000013159
|
LOC680121
|
similar to heat shock protein 8 |
| chr3_-_165537940 | 0.01 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr5_-_124403195 | 0.01 |
ENSRNOT00000067850
|
C8a
|
complement C8 alpha chain |
| chr3_+_307204 | 0.01 |
ENSRNOT00000062080
|
Nxph2
|
neurexophilin 2 |
| chr20_+_26988774 | 0.01 |
ENSRNOT00000090083
|
Mypn
|
myopalladin |
| chr7_-_144837583 | 0.01 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr10_-_86930947 | 0.01 |
ENSRNOT00000081440
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr17_+_9736786 | 0.01 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chrX_-_15467875 | 0.01 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chrM_-_14061 | 0.01 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr7_-_34406318 | 0.01 |
ENSRNOT00000007331
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr1_+_280383579 | 0.01 |
ENSRNOT00000064022
ENSRNOT00000042977 |
Kcnk18
|
potassium two pore domain channel subfamily K member 18 |
| chr4_-_10269748 | 0.00 |
ENSRNOT00000074662
|
Fam185a
|
family with sequence similarity 185, member A |
| chr1_+_266255797 | 0.00 |
ENSRNOT00000027047
|
Trim8
|
tripartite motif-containing 8 |
| chr1_+_260289589 | 0.00 |
ENSRNOT00000051058
|
Dntt
|
DNA nucleotidylexotransferase |
| chr9_-_5330815 | 0.00 |
ENSRNOT00000014548
|
Slc5a7
|
solute carrier family 5 member 7 |
| chr8_-_1450138 | 0.00 |
ENSRNOT00000008062
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr6_+_22696397 | 0.00 |
ENSRNOT00000011630
|
Alk
|
anaplastic lymphoma receptor tyrosine kinase |
| chr6_-_67085390 | 0.00 |
ENSRNOT00000083277
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr14_-_44225713 | 0.00 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr11_-_35749464 | 0.00 |
ENSRNOT00000078818
ENSRNOT00000078425 |
Erg
|
ERG, ETS transcription factor |
| chr5_+_57845819 | 0.00 |
ENSRNOT00000017712
|
Nudt2
|
nudix hydrolase 2 |
| chr14_-_64476796 | 0.00 |
ENSRNOT00000029104
|
Gba3
|
glucosidase, beta, acid 3 |
| chrX_+_124894466 | 0.00 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr2_-_258932200 | 0.00 |
ENSRNOT00000045905
|
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chrX_-_72370044 | 0.00 |
ENSRNOT00000004224
|
Hdac8
|
histone deacetylase 8 |
| chr3_-_2444281 | 0.00 |
ENSRNOT00000013863
|
Tubb4b
|
tubulin, beta 4B class IVb |
| chr7_-_102298522 | 0.00 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr10_+_84135116 | 0.00 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chr15_-_33752665 | 0.00 |
ENSRNOT00000034102
|
Zfhx2
|
zinc finger homeobox 2 |
| chr2_-_95472115 | 0.00 |
ENSRNOT00000015720
|
Stmn2
|
stathmin 2 |
| chr11_-_17340373 | 0.00 |
ENSRNOT00000089762
|
AABR07033324.1
|
|
| chr2_-_252451999 | 0.00 |
ENSRNOT00000021861
|
Dnase2b
|
deoxyribonuclease 2 beta |
| chr8_-_46675544 | 0.00 |
ENSRNOT00000052293
|
Tecta
|
tectorin alpha |
| chr5_-_28164326 | 0.00 |
ENSRNOT00000088165
|
Slc26a7
|
solute carrier family 26 member 7 |
| chr1_+_214375515 | 0.00 |
ENSRNOT00000024863
|
Taldo1
|
transaldolase 1 |
| chr3_-_81304181 | 0.00 |
ENSRNOT00000079746
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr20_+_25990304 | 0.00 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr10_+_65767053 | 0.00 |
ENSRNOT00000078897
|
Vtn
|
vitronectin |
| chrX_-_77675487 | 0.00 |
ENSRNOT00000042799
|
Cysltr1
|
cysteinyl leukotriene receptor 1 |
| chr18_-_77579969 | 0.00 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr1_+_1897338 | 0.00 |
ENSRNOT00000021437
|
Ppil4
|
peptidylprolyl isomerase like 4 |
| chrM_+_14136 | 0.00 |
ENSRNOT00000042098
|
Mt-cyb
|
mitochondrially encoded cytochrome b |
| chr2_+_22909569 | 0.00 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr14_-_23604834 | 0.00 |
ENSRNOT00000002760
|
Stap1
|
signal transducing adaptor family member 1 |
| chr7_-_119689938 | 0.00 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr2_-_235177275 | 0.00 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr10_+_89286047 | 0.00 |
ENSRNOT00000085831
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr2_-_238246931 | 0.00 |
ENSRNOT00000086303
|
Gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr1_+_83714347 | 0.00 |
ENSRNOT00000085245
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr6_+_136004214 | 0.00 |
ENSRNOT00000013695
|
NEWGENE_619861
|
eukaryotic translation initiation factor 5 |
| chr15_+_44799334 | 0.00 |
ENSRNOT00000018599
|
Nefl
|
neurofilament light |
| chr8_+_117068582 | 0.00 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr6_-_72786830 | 0.00 |
ENSRNOT00000009020
|
Dtd2
|
D-tyrosyl-tRNA deacylase 2 |
| chr2_+_22910236 | 0.00 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr2_+_68820615 | 0.00 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr2_+_143475323 | 0.00 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr7_+_34533543 | 0.00 |
ENSRNOT00000007412
ENSRNOT00000084185 |
Ntn4
|
netrin 4 |
| chr5_-_62621737 | 0.00 |
ENSRNOT00000011573
|
Gabbr2
|
gamma-aminobutyric acid type B receptor subunit 2 |
| chr6_+_109043880 | 0.00 |
ENSRNOT00000008730
|
Eif2b2
|
eukaryotic translation initiation factor 2B subunit beta |
| chr10_+_89285855 | 0.00 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |