Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
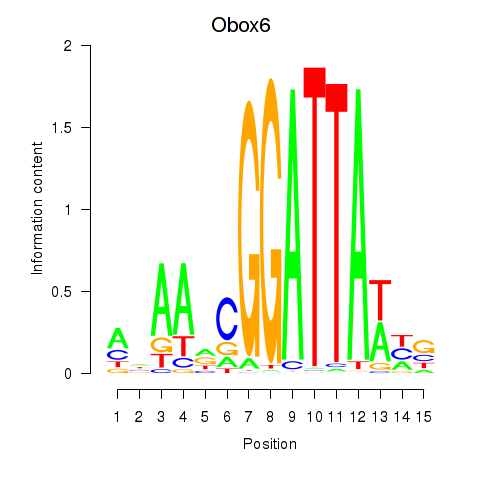
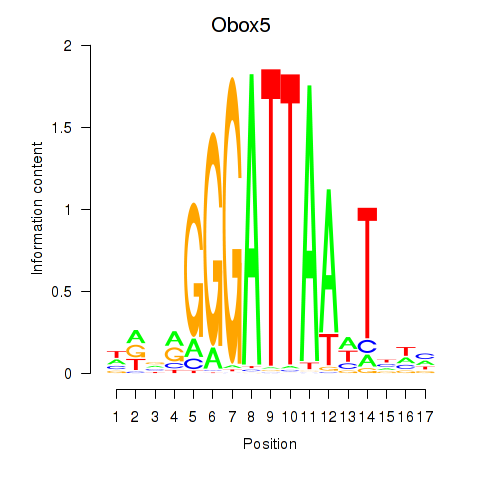
Results for Obox6_Obox5
Z-value: 0.92
Transcription factors associated with Obox6_Obox5
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Obox6_Obox5 motif
Sorted Z-values of Obox6_Obox5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_71973419 | 0.57 |
ENSRNOT00000079895
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr7_-_93826665 | 0.53 |
ENSRNOT00000011344
|
Tnfrsf11b
|
TNF receptor superfamily member 11B |
| chr14_+_9226125 | 0.50 |
ENSRNOT00000088047
|
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr1_-_97785490 | 0.48 |
ENSRNOT00000019364
|
LOC499136
|
LRRGT00021 |
| chr7_-_116106368 | 0.42 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr6_+_106084815 | 0.39 |
ENSRNOT00000058195
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chrX_-_70460536 | 0.38 |
ENSRNOT00000076824
|
Pdzd11
|
PDZ domain containing 11 |
| chr7_-_116063078 | 0.38 |
ENSRNOT00000076932
ENSRNOT00000035496 |
Gml
|
glycosylphosphatidylinositol anchored molecule like |
| chr1_+_87563975 | 0.36 |
ENSRNOT00000088772
|
Zfp30
|
zinc finger protein 30 |
| chr3_+_124896618 | 0.36 |
ENSRNOT00000028888
|
Cds2
|
CDP-diacylglycerol synthase 2 |
| chr13_-_49828720 | 0.36 |
ENSRNOT00000012984
|
Mdm4
|
MDM4, p53 regulator |
| chr2_-_167607919 | 0.35 |
ENSRNOT00000089083
|
AABR07011733.1
|
|
| chr20_+_28572242 | 0.35 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr11_+_71151132 | 0.33 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr11_+_57207656 | 0.33 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr4_+_66090209 | 0.31 |
ENSRNOT00000031193
|
Ttc26
|
tetratricopeptide repeat domain 26 |
| chr6_-_107325345 | 0.30 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr1_+_84685931 | 0.30 |
ENSRNOT00000063839
|
Zfp780b
|
zinc finger protein 780B |
| chr19_+_27603307 | 0.30 |
ENSRNOT00000074702
|
AABR07043347.1
|
|
| chr1_-_4011161 | 0.29 |
ENSRNOT00000044778
|
Stxbp5
|
syntaxin binding protein 5 |
| chr17_+_35396286 | 0.29 |
ENSRNOT00000089613
ENSRNOT00000092638 |
Exoc2
|
exocyst complex component 2 |
| chr13_+_50103189 | 0.29 |
ENSRNOT00000004078
|
Atp2b4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chrM_+_11736 | 0.28 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr12_-_10335499 | 0.28 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr6_+_135610743 | 0.27 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr7_-_118474609 | 0.27 |
ENSRNOT00000089076
|
AABR07058464.1
|
|
| chr20_-_8574082 | 0.26 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr9_-_17209220 | 0.25 |
ENSRNOT00000083764
|
Gtpbp2
|
GTP binding protein 2 |
| chr20_-_5927070 | 0.25 |
ENSRNOT00000059264
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr18_-_74485139 | 0.25 |
ENSRNOT00000022598
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr7_-_124929025 | 0.25 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr5_+_6373583 | 0.25 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr6_+_144156175 | 0.25 |
ENSRNOT00000006582
|
Esyt2
|
extended synaptotagmin 2 |
| chr4_+_152450126 | 0.24 |
ENSRNOT00000076930
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr9_-_88816898 | 0.24 |
ENSRNOT00000083667
|
Slc19a3
|
solute carrier family 19 member 3 |
| chr13_+_34365147 | 0.23 |
ENSRNOT00000093066
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr14_+_39663421 | 0.23 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr1_-_4011511 | 0.23 |
ENSRNOT00000040559
|
Stxbp5
|
syntaxin binding protein 5 |
| chr2_+_66940057 | 0.22 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr4_+_163112301 | 0.22 |
ENSRNOT00000087113
|
Clec12a
|
C-type lectin domain family 12, member A |
| chr1_+_99505677 | 0.22 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr7_-_120770435 | 0.22 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr8_-_69466618 | 0.22 |
ENSRNOT00000042925
|
Ids
|
iduronate 2-sulfatase |
| chr17_-_32783427 | 0.22 |
ENSRNOT00000059921
|
Serpinb6b
|
serine (or cysteine) peptidase inhibitor, clade B, member 6b |
| chr7_+_20262680 | 0.21 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr5_+_5616483 | 0.21 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr3_+_48106099 | 0.20 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr2_+_34186091 | 0.20 |
ENSRNOT00000016129
|
Sgtb
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr8_-_22150005 | 0.20 |
ENSRNOT00000041678
|
Tyk2
|
tyrosine kinase 2 |
| chr13_-_113817995 | 0.20 |
ENSRNOT00000057151
|
Cd46
|
CD46 molecule |
| chr10_+_10875983 | 0.20 |
ENSRNOT00000052265
|
LOC679823
|
similar to 60S ribosomal protein L37a |
| chr12_-_19294888 | 0.19 |
ENSRNOT00000001815
|
Zfp113
|
zinc finger protein 3 |
| chr19_-_56789404 | 0.19 |
ENSRNOT00000058652
|
RGD1563991
|
similar to TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr1_+_192379543 | 0.19 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr4_-_51586881 | 0.19 |
ENSRNOT00000064983
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chr8_-_116532169 | 0.18 |
ENSRNOT00000085364
|
Rbm5
|
RNA binding motif protein 5 |
| chr3_+_94530586 | 0.18 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr11_+_61531416 | 0.18 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr10_-_55997299 | 0.18 |
ENSRNOT00000074505
|
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr3_-_121488540 | 0.18 |
ENSRNOT00000023845
|
Zc3h8
|
zinc finger CCCH type containing 8 |
| chr4_+_140092352 | 0.17 |
ENSRNOT00000008892
|
Setmar
|
SET domain and mariner transposase fusion gene |
| chr2_+_127489771 | 0.17 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr12_-_10391270 | 0.17 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr6_-_54059119 | 0.17 |
ENSRNOT00000005521
|
Hdac9
|
histone deacetylase 9 |
| chr17_+_19249952 | 0.17 |
ENSRNOT00000023140
|
Atxn1
|
ataxin 1 |
| chr1_+_11963836 | 0.17 |
ENSRNOT00000074325
|
AABR07000398.1
|
|
| chr15_+_36809361 | 0.17 |
ENSRNOT00000076667
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr10_+_56381813 | 0.17 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr11_+_30550141 | 0.16 |
ENSRNOT00000002866
|
Hunk
|
hormonally upregulated Neu-associated kinase |
| chr6_-_115616766 | 0.16 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chrM_+_9870 | 0.16 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr3_-_162059524 | 0.16 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr1_+_65522118 | 0.16 |
ENSRNOT00000058563
|
Mzf1
|
myeloid zinc finger 1 |
| chr3_+_154863072 | 0.16 |
ENSRNOT00000055200
|
Snhg11
|
small nucleolar RNA host gene 11 |
| chr17_-_67904674 | 0.16 |
ENSRNOT00000078532
|
Klf6
|
Kruppel-like factor 6 |
| chr12_-_15996958 | 0.16 |
ENSRNOT00000039726
|
Amz1
|
archaelysin family metallopeptidase 1 |
| chr18_-_71701423 | 0.16 |
ENSRNOT00000024716
|
Ctif
|
cap binding complex dependent translation initiation factor |
| chr2_-_2779109 | 0.15 |
ENSRNOT00000017098
|
Rfesd
|
Rieske (Fe-S) domain containing |
| chr2_+_206454208 | 0.15 |
ENSRNOT00000026863
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr2_-_106703401 | 0.15 |
ENSRNOT00000043933
|
AABR07009638.1
|
|
| chr4_+_9981958 | 0.15 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr17_+_16455026 | 0.15 |
ENSRNOT00000022932
|
Zfp169
|
zinc finger protein 169 |
| chr14_+_108415068 | 0.15 |
ENSRNOT00000083763
|
Pus10
|
pseudouridylate synthase 10 |
| chr7_+_70946228 | 0.15 |
ENSRNOT00000039306
|
Stat6
|
signal transducer and activator of transcription 6 |
| chr13_-_70625842 | 0.15 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr14_+_8182383 | 0.15 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr10_+_35343189 | 0.14 |
ENSRNOT00000083688
|
Mapk9
|
mitogen-activated protein kinase 9 |
| chr4_-_150616895 | 0.14 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr1_+_15782477 | 0.14 |
ENSRNOT00000088025
ENSRNOT00000089966 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr10_-_35175647 | 0.14 |
ENSRNOT00000078960
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr6_+_86862790 | 0.14 |
ENSRNOT00000093492
|
Fancm
|
Fanconi anemia, complementation group M |
| chr8_+_21611319 | 0.14 |
ENSRNOT00000068461
|
Zfp846
|
zinc finger protein 846 |
| chr6_+_122729874 | 0.14 |
ENSRNOT00000039509
ENSRNOT00000040440 |
Zc3h14
|
zinc finger CCCH type containing 14 |
| chr16_-_14360555 | 0.14 |
ENSRNOT00000065460
|
LOC290595
|
hypothetical gene supported by AF152002 |
| chr4_+_87026530 | 0.14 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr3_+_63379031 | 0.13 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr18_+_30864216 | 0.13 |
ENSRNOT00000027015
|
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr6_+_80159364 | 0.13 |
ENSRNOT00000005439
|
Pnn
|
pinin, desmosome associated protein |
| chr1_+_141447022 | 0.13 |
ENSRNOT00000020690
|
AC096024.1
|
|
| chr9_+_12114977 | 0.13 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr4_+_181481147 | 0.13 |
ENSRNOT00000002523
|
Klhl42
|
kelch-like family, member 42 |
| chr13_-_70980913 | 0.13 |
ENSRNOT00000003713
|
Npl
|
N-acetylneuraminate pyruvate lyase |
| chr2_+_144646308 | 0.13 |
ENSRNOT00000078337
ENSRNOT00000093407 |
Dclk1
|
doublecortin-like kinase 1 |
| chr6_+_27328406 | 0.13 |
ENSRNOT00000091159
ENSRNOT00000044278 |
Otof
|
otoferlin |
| chr18_-_32207749 | 0.13 |
ENSRNOT00000090882
ENSRNOT00000018843 |
Arhgap26
|
Rho GTPase activating protein 26 |
| chr6_-_4520604 | 0.12 |
ENSRNOT00000042230
ENSRNOT00000043870 ENSRNOT00000070918 ENSRNOT00000046246 ENSRNOT00000052367 ENSRNOT00000042251 |
Slc8a1
|
solute carrier family 8 member A1 |
| chr7_+_27620458 | 0.12 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr4_+_170518673 | 0.12 |
ENSRNOT00000011803
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr15_+_41448064 | 0.12 |
ENSRNOT00000019551
|
Sacs
|
sacsin molecular chaperone |
| chr3_-_165700489 | 0.12 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr6_+_9790422 | 0.12 |
ENSRNOT00000020959
|
Prkce
|
protein kinase C, epsilon |
| chr18_+_56414488 | 0.12 |
ENSRNOT00000088988
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr1_+_234252757 | 0.12 |
ENSRNOT00000091814
|
Rorb
|
RAR-related orphan receptor B |
| chr14_-_114649173 | 0.12 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr13_+_77602249 | 0.12 |
ENSRNOT00000003407
ENSRNOT00000076589 |
Tnr
|
tenascin R |
| chr8_-_96516975 | 0.12 |
ENSRNOT00000067996
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr4_-_26255639 | 0.12 |
ENSRNOT00000031118
|
AC126960.1
|
|
| chrX_+_128416722 | 0.12 |
ENSRNOT00000009336
ENSRNOT00000085110 |
Xiap
|
X-linked inhibitor of apoptosis |
| chr8_-_84506328 | 0.12 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr14_-_46153212 | 0.12 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr2_+_248178389 | 0.12 |
ENSRNOT00000037339
|
Gbp5
|
guanylate binding protein 5 |
| chr3_-_80012750 | 0.12 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr12_-_41448668 | 0.11 |
ENSRNOT00000001856
|
Rasal1
|
RAS protein activator like 1 (GAP1 like) |
| chr10_+_5930298 | 0.11 |
ENSRNOT00000044626
|
Grin2a
|
glutamate ionotropic receptor NMDA type subunit 2A |
| chr6_-_91581262 | 0.11 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr13_+_6679368 | 0.11 |
ENSRNOT00000067198
ENSRNOT00000092965 |
Cntnap5c
|
contactin associated protein-like 5C |
| chr3_+_97723901 | 0.11 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr1_-_84732242 | 0.11 |
ENSRNOT00000025586
|
LOC102553760
|
zinc finger protein 59-like |
| chr9_+_73687743 | 0.11 |
ENSRNOT00000079495
ENSRNOT00000017197 |
Rpe
|
ribulose-5-phosphate-3-epimerase |
| chr17_+_87274944 | 0.11 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr4_-_153593773 | 0.11 |
ENSRNOT00000082982
|
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr13_-_100450209 | 0.11 |
ENSRNOT00000090712
|
Lbr
|
lamin B receptor |
| chr8_+_93439648 | 0.11 |
ENSRNOT00000043008
|
LOC100359563
|
ribosomal protein S20-like |
| chr4_-_85329362 | 0.11 |
ENSRNOT00000014925
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr3_+_98297554 | 0.11 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr10_-_67751002 | 0.11 |
ENSRNOT00000000254
|
RGD1310429
|
similar to Protein Njmu-R1 |
| chr4_-_148446303 | 0.11 |
ENSRNOT00000017633
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr10_-_62698541 | 0.11 |
ENSRNOT00000019759
|
Coro6
|
coronin 6 |
| chr3_+_56030915 | 0.11 |
ENSRNOT00000010489
|
Phospho2
|
phosphatase, orphan 2 |
| chr7_-_129919946 | 0.11 |
ENSRNOT00000079976
|
AABR07058658.1
|
|
| chr1_+_78893271 | 0.11 |
ENSRNOT00000029825
|
Pnmal1
|
paraneoplastic Ma antigen family-like 1 |
| chr12_-_30770791 | 0.11 |
ENSRNOT00000093734
|
Sfswap
|
splicing factor SWAP homolog |
| chr13_-_73056765 | 0.11 |
ENSRNOT00000000049
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr17_+_54298409 | 0.11 |
ENSRNOT00000084919
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr5_-_161879133 | 0.11 |
ENSRNOT00000041175
|
Prdm2
|
PR/SET domain 2 |
| chr2_+_54191538 | 0.11 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr7_+_124680294 | 0.10 |
ENSRNOT00000014720
|
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr1_-_211196868 | 0.10 |
ENSRNOT00000022714
|
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr2_-_178521038 | 0.10 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr7_+_13151592 | 0.10 |
ENSRNOT00000010203
|
Vom2r53
|
vomeronasal 2 receptor, 53 |
| chr16_+_84465656 | 0.10 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr19_-_22194740 | 0.10 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr11_-_38457373 | 0.10 |
ENSRNOT00000041177
|
Zfp295
|
zinc finger protein 295 |
| chr20_+_20236151 | 0.10 |
ENSRNOT00000079630
|
Ank3
|
ankyrin 3 |
| chr14_+_109533792 | 0.10 |
ENSRNOT00000067690
|
LOC108348154
|
ankyrin repeat domain-containing protein 29-like |
| chr9_-_30844199 | 0.10 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr18_-_86878142 | 0.10 |
ENSRNOT00000058139
|
Dok6
|
docking protein 6 |
| chr5_-_59085676 | 0.10 |
ENSRNOT00000068105
|
Msmp
|
microseminoprotein, prostate associated |
| chr11_-_77593171 | 0.10 |
ENSRNOT00000002645
ENSRNOT00000043498 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr12_-_38638536 | 0.10 |
ENSRNOT00000001690
|
Mlxip
|
MLX interacting protein |
| chr6_-_50923510 | 0.10 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr6_+_78567970 | 0.10 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr2_+_86891092 | 0.10 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr16_+_46731403 | 0.10 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr16_-_21362955 | 0.10 |
ENSRNOT00000039607
|
Gmip
|
Gem-interacting protein |
| chr17_+_69654170 | 0.10 |
ENSRNOT00000058351
|
Akr1c19
|
aldo-keto reductase family 1, member C19 |
| chr11_+_66316606 | 0.10 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr6_-_38228379 | 0.10 |
ENSRNOT00000084924
|
Mycn
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr4_-_15859132 | 0.10 |
ENSRNOT00000082161
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr4_-_155763500 | 0.10 |
ENSRNOT00000071745
|
LOC100909595
|
solute carrier family 2, facilitated glucose transporter member 3-like |
| chr4_+_179398621 | 0.10 |
ENSRNOT00000049474
ENSRNOT00000067506 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr1_+_224998172 | 0.10 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr14_+_83298674 | 0.09 |
ENSRNOT00000024813
|
Pisd
|
phosphatidylserine decarboxylase |
| chr2_+_58448917 | 0.09 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr18_+_30831365 | 0.09 |
ENSRNOT00000067185
|
Pcdhga4
|
protocadherin gamma subfamily A, 4 |
| chr3_-_79282493 | 0.09 |
ENSRNOT00000049832
|
Ptprj
|
protein tyrosine phosphatase, receptor type, J |
| chr1_+_104106245 | 0.09 |
ENSRNOT00000019175
ENSRNOT00000081660 |
Zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr10_+_11786121 | 0.09 |
ENSRNOT00000033919
|
Slx4
|
SLX4 structure-specific endonuclease subunit |
| chr11_+_60102121 | 0.09 |
ENSRNOT00000045521
|
Tmprss7
|
transmembrane protease, serine 7 |
| chr3_-_12155098 | 0.09 |
ENSRNOT00000082696
|
Garnl3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr13_-_89545182 | 0.09 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr1_+_72874404 | 0.09 |
ENSRNOT00000058900
|
Dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr9_-_81864202 | 0.09 |
ENSRNOT00000084964
|
Zfp142
|
zinc finger protein 142 |
| chr14_+_81858737 | 0.09 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr10_-_2037891 | 0.09 |
ENSRNOT00000004563
|
Ercc4
|
ERCC excision repair 4, endonuclease catalytic subunit |
| chr4_-_117589464 | 0.09 |
ENSRNOT00000021167
|
Nat8f1
|
N-acetyltransferase 8 (GCN5-related) family member 1 |
| chr1_-_4011353 | 0.09 |
ENSRNOT00000049699
|
Stxbp5
|
syntaxin binding protein 5 |
| chr1_-_248376750 | 0.09 |
ENSRNOT00000089073
|
Gldc
|
glycine decarboxylase |
| chr18_+_30487264 | 0.09 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr6_-_1454480 | 0.09 |
ENSRNOT00000072810
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr18_-_76653056 | 0.09 |
ENSRNOT00000078496
|
Adnp2
|
ADNP homeobox 2 |
| chr3_+_80833272 | 0.09 |
ENSRNOT00000023583
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr3_+_11392791 | 0.09 |
ENSRNOT00000064792
|
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr8_+_131965134 | 0.09 |
ENSRNOT00000078308
|
Znf660
|
zinc finger protein 660 |
| chr4_+_147455533 | 0.09 |
ENSRNOT00000081957
|
Tsen2
|
tRNA splicing endonuclease subunit 2 |
| chr8_-_94920441 | 0.09 |
ENSRNOT00000014165
|
Cep162
|
centrosomal protein 162 |
| chr14_+_113867209 | 0.09 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr19_-_15540773 | 0.09 |
ENSRNOT00000022359
|
Lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr4_+_85427555 | 0.09 |
ENSRNOT00000015469
|
Fam188b
|
family with sequence similarity 188, member B |
| chr3_-_21669957 | 0.09 |
ENSRNOT00000012354
ENSRNOT00000078718 |
Rc3h2
|
ring finger and CCCH-type domains 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox6_Obox5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.1 | 0.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.3 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.3 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 0.4 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:2000293 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.2 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.4 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0042938 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 0.0 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) |
| 0.0 | 0.1 | GO:0061591 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:0046548 | amacrine cell differentiation(GO:0035881) retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.0 | GO:0042496 | detection of diacyl bacterial lipopeptide(GO:0042496) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.1 | GO:1990910 | response to hypobaric hypoxia(GO:1990910) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.2 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.1 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.2 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 0.4 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.2 | GO:0015563 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.0 | 0.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |