Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
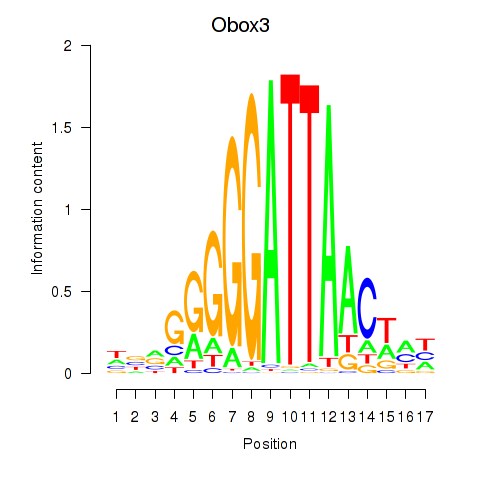
Results for Obox3
Z-value: 0.16
Transcription factors associated with Obox3
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Obox3 motif
Sorted Z-values of Obox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_260596777 | 0.07 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr1_+_89220083 | 0.04 |
ENSRNOT00000093144
|
Dmkn
|
dermokine |
| chr1_+_72661211 | 0.03 |
ENSRNOT00000033197
|
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr4_+_123586231 | 0.02 |
ENSRNOT00000049657
ENSRNOT00000013232 |
Grip2
|
glutamate receptor interacting protein 2 |
| chr8_-_87315955 | 0.02 |
ENSRNOT00000081437
|
Filip1
|
filamin A interacting protein 1 |
| chr5_+_138300107 | 0.02 |
ENSRNOT00000047151
|
Cldn19
|
claudin 19 |
| chr19_+_60017746 | 0.02 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr4_+_117962319 | 0.02 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr5_+_48303366 | 0.02 |
ENSRNOT00000009973
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr5_-_115387377 | 0.02 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr1_-_266074181 | 0.02 |
ENSRNOT00000026378
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr3_+_112242270 | 0.01 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr6_-_132958546 | 0.01 |
ENSRNOT00000041903
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr8_-_107490093 | 0.01 |
ENSRNOT00000046832
|
LOC684466
|
similar to Fas apoptotic inhibitory molecule 1 (rFAIM) |
| chr10_-_55560422 | 0.01 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr12_-_22477052 | 0.01 |
ENSRNOT00000075504
|
Ache
|
acetylcholinesterase |
| chr10_-_103481153 | 0.01 |
ENSRNOT00000035639
|
Cd300lb
|
CD300 molecule-like family member b |
| chr3_+_43255567 | 0.01 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr18_-_37776453 | 0.01 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr2_+_208738132 | 0.01 |
ENSRNOT00000023972
|
AABR07012795.1
|
|
| chr19_+_51985170 | 0.01 |
ENSRNOT00000019443
|
Hsbp1
|
heat shock factor binding protein 1 |
| chr1_-_175796040 | 0.01 |
ENSRNOT00000024060
|
Mrvi1
|
murine retrovirus integration site 1 homolog |
| chrX_-_116638508 | 0.01 |
ENSRNOT00000030400
|
Lhfpl1
|
lipoma HMGIC fusion partner-like 1 |
| chr5_-_61734011 | 0.01 |
ENSRNOT00000012915
|
Tstd2
|
thiosulfate sulfurtransferase like domain containing 2 |
| chr13_+_77940454 | 0.01 |
ENSRNOT00000003460
|
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr10_-_91807937 | 0.01 |
ENSRNOT00000005074
|
Wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr15_-_45524582 | 0.01 |
ENSRNOT00000081912
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr9_+_10044756 | 0.01 |
ENSRNOT00000071188
|
Pspn
|
persephin |
| chr5_-_162602339 | 0.01 |
ENSRNOT00000071779
|
RGD1563334
|
similar to novel protein similar to esterases |
| chr10_+_54512983 | 0.01 |
ENSRNOT00000005204
|
Stx8
|
syntaxin 8 |
| chr2_+_93712992 | 0.01 |
ENSRNOT00000059326
|
Fabp12
|
fatty acid binding protein 12 |
| chrM_+_7758 | 0.01 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chrM_+_7919 | 0.00 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr4_-_100783750 | 0.00 |
ENSRNOT00000078956
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr7_+_94795214 | 0.00 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr7_-_49741540 | 0.00 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr18_+_40962146 | 0.00 |
ENSRNOT00000035656
|
Arl14epl
|
ADP ribosylation factor like GTPase 14 effector protein like |
| chr1_+_213676954 | 0.00 |
ENSRNOT00000050551
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr8_-_116465801 | 0.00 |
ENSRNOT00000085915
|
Sema3f
|
semaphorin 3F |
| chr2_-_218658761 | 0.00 |
ENSRNOT00000018318
|
S1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr10_+_56605140 | 0.00 |
ENSRNOT00000024079
|
Phf23
|
PHD finger protein 23 |
| chr7_-_27213651 | 0.00 |
ENSRNOT00000077041
ENSRNOT00000076468 |
Tdg
|
thymine-DNA glycosylase |
| chr10_-_55851235 | 0.00 |
ENSRNOT00000010790
|
Gucy2d
|
guanylate cyclase 2D, retinal |
| chr18_+_56544652 | 0.00 |
ENSRNOT00000024171
|
Pde6a
|
phosphodiesterase 6A |
| chr6_-_26855658 | 0.00 |
ENSRNOT00000011635
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr8_+_94686938 | 0.00 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr5_+_159484370 | 0.00 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr5_+_61734118 | 0.00 |
ENSRNOT00000030825
|
Ncbp1
|
nuclear cap binding protein subunit 1 |
| chr1_+_213686046 | 0.00 |
ENSRNOT00000019808
|
LOC108348167
|
NACHT, LRR and PYD domains-containing protein 6-like |
| chr10_+_83954279 | 0.00 |
ENSRNOT00000006594
|
Ttll6
|
tubulin tyrosine ligase like 6 |
| chr5_+_81431600 | 0.00 |
ENSRNOT00000013675
|
Trim32
|
tripartite motif-containing 32 |
| chr5_-_77031120 | 0.00 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chr7_-_27214236 | 0.00 |
ENSRNOT00000034213
|
Tdg
|
thymine-DNA glycosylase |
| chr10_-_15305549 | 0.00 |
ENSRNOT00000027353
|
Pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr19_+_9622611 | 0.00 |
ENSRNOT00000061498
|
Slc38a7
|
solute carrier family 38, member 7 |
| chr1_-_22404002 | 0.00 |
ENSRNOT00000044098
|
Taar8a
|
trace amine-associated receptor 8a |
| chr2_+_187988925 | 0.00 |
ENSRNOT00000093033
ENSRNOT00000093016 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr11_-_80981415 | 0.00 |
ENSRNOT00000002499
ENSRNOT00000002496 |
St6gal1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr5_+_98387291 | 0.00 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr10_+_89459650 | 0.00 |
ENSRNOT00000028149
ENSRNOT00000077942 |
Nbr1
|
NBR1, autophagy cargo receptor |
| chr2_+_60966789 | 0.00 |
ENSRNOT00000025490
|
Slc45a2
|
solute carrier family 45, member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox3
Gene Ontology Analysis
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |