Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
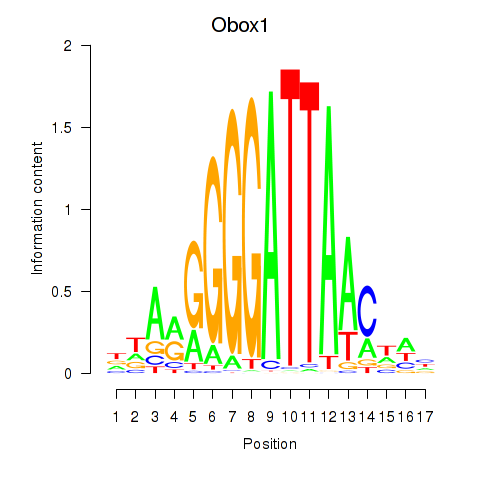
Results for Obox1
Z-value: 0.41
Transcription factors associated with Obox1
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Obox1 motif
Sorted Z-values of Obox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_70460536 | 0.16 |
ENSRNOT00000076824
|
Pdzd11
|
PDZ domain containing 11 |
| chr3_+_124896618 | 0.15 |
ENSRNOT00000028888
|
Cds2
|
CDP-diacylglycerol synthase 2 |
| chr2_-_167607919 | 0.15 |
ENSRNOT00000089083
|
AABR07011733.1
|
|
| chr13_+_34365147 | 0.14 |
ENSRNOT00000093066
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr5_+_6373583 | 0.13 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chrM_+_9870 | 0.12 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr14_-_71973419 | 0.12 |
ENSRNOT00000079895
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr6_+_144156175 | 0.11 |
ENSRNOT00000006582
|
Esyt2
|
extended synaptotagmin 2 |
| chr1_+_11963836 | 0.11 |
ENSRNOT00000074325
|
AABR07000398.1
|
|
| chr1_+_65851060 | 0.10 |
ENSRNOT00000036880
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr1_+_89220083 | 0.10 |
ENSRNOT00000093144
|
Dmkn
|
dermokine |
| chr9_-_94634109 | 0.10 |
ENSRNOT00000081131
|
AABR07068274.1
|
|
| chr4_+_163112301 | 0.10 |
ENSRNOT00000087113
|
Clec12a
|
C-type lectin domain family 12, member A |
| chr2_+_127489771 | 0.10 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr15_+_36865548 | 0.10 |
ENSRNOT00000076460
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr1_-_97785490 | 0.09 |
ENSRNOT00000019364
|
LOC499136
|
LRRGT00021 |
| chr1_-_219438779 | 0.09 |
ENSRNOT00000029237
|
Tbc1d10c
|
TBC1 domain family, member 10C |
| chr20_-_5927070 | 0.09 |
ENSRNOT00000059264
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr7_-_116063078 | 0.09 |
ENSRNOT00000076932
ENSRNOT00000035496 |
Gml
|
glycosylphosphatidylinositol anchored molecule like |
| chrX_+_152885246 | 0.09 |
ENSRNOT00000087074
|
AABR07042326.3
|
|
| chr6_+_106084815 | 0.09 |
ENSRNOT00000058195
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr4_-_51586881 | 0.09 |
ENSRNOT00000064983
|
Iqub
|
IQ motif and ubiquitin domain containing |
| chr1_-_101664436 | 0.09 |
ENSRNOT00000028522
|
Sec1
|
secretory blood group 1 |
| chr10_+_10875983 | 0.08 |
ENSRNOT00000052265
|
LOC679823
|
similar to 60S ribosomal protein L37a |
| chr7_+_94795214 | 0.08 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr2_+_66940057 | 0.08 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chrM_+_11736 | 0.08 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr9_+_12114977 | 0.08 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr18_-_74485139 | 0.08 |
ENSRNOT00000022598
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr16_+_39144972 | 0.07 |
ENSRNOT00000086728
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr4_-_85329362 | 0.07 |
ENSRNOT00000014925
|
Crhr2
|
corticotropin releasing hormone receptor 2 |
| chr5_-_173611202 | 0.07 |
ENSRNOT00000047854
|
Agrn
|
agrin |
| chr1_+_72874404 | 0.07 |
ENSRNOT00000058900
|
Dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr17_+_16455026 | 0.07 |
ENSRNOT00000022932
|
Zfp169
|
zinc finger protein 169 |
| chr6_-_91581262 | 0.07 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr2_-_260596777 | 0.07 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr9_+_49903085 | 0.06 |
ENSRNOT00000022601
|
RGD1310553
|
similar to expressed sequence AI597479 |
| chr13_-_49828720 | 0.06 |
ENSRNOT00000012984
|
Mdm4
|
MDM4, p53 regulator |
| chr3_+_97723901 | 0.06 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr1_-_167884690 | 0.06 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr11_-_77593171 | 0.06 |
ENSRNOT00000002645
ENSRNOT00000043498 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr5_-_9094616 | 0.06 |
ENSRNOT00000072210
|
Sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr17_+_2165906 | 0.06 |
ENSRNOT00000047043
|
RGD1563581
|
similar to S100 calcium binding protein A11 (calizzarin) |
| chr20_+_31102476 | 0.06 |
ENSRNOT00000078719
|
Lrrc20
|
leucine rich repeat containing 20 |
| chr20_+_2057878 | 0.06 |
ENSRNOT00000051480
|
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr14_+_71542057 | 0.06 |
ENSRNOT00000082592
ENSRNOT00000083701 ENSRNOT00000084322 |
Prom1
|
prominin 1 |
| chr2_-_2779109 | 0.06 |
ENSRNOT00000017098
|
Rfesd
|
Rieske (Fe-S) domain containing |
| chr3_+_114355798 | 0.05 |
ENSRNOT00000024658
ENSRNOT00000036435 |
Slc28a2
|
solute carrier family 28 member 2 |
| chr6_-_50941248 | 0.05 |
ENSRNOT00000084533
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr19_+_60017746 | 0.05 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr17_+_69634890 | 0.05 |
ENSRNOT00000029049
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr11_-_69316848 | 0.05 |
ENSRNOT00000033420
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr2_-_154542557 | 0.05 |
ENSRNOT00000013392
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr1_+_141447022 | 0.05 |
ENSRNOT00000020690
|
AC096024.1
|
|
| chr6_-_42630983 | 0.05 |
ENSRNOT00000071977
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr7_+_123482255 | 0.05 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr7_+_71004417 | 0.05 |
ENSRNOT00000005575
|
Myo1a
|
myosin IA |
| chr1_-_67302751 | 0.05 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr4_-_23135354 | 0.05 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr9_+_73334618 | 0.05 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr4_-_70934295 | 0.05 |
ENSRNOT00000020616
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr11_-_87434482 | 0.05 |
ENSRNOT00000074346
ENSRNOT00000002557 |
Lrrc74b
|
leucine rich repeat containing 74B |
| chr3_+_80833272 | 0.05 |
ENSRNOT00000023583
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr13_-_70625842 | 0.05 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr3_-_165700489 | 0.05 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr7_-_117732339 | 0.05 |
ENSRNOT00000092917
ENSRNOT00000020696 |
Foxh1
|
forkhead box H1 |
| chr3_-_175479034 | 0.04 |
ENSRNOT00000086761
|
Hrh3
|
histamine receptor H3 |
| chr4_-_30276372 | 0.04 |
ENSRNOT00000011823
|
Pon1
|
paraoxonase 1 |
| chr4_-_153593773 | 0.04 |
ENSRNOT00000082982
|
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr2_-_235249571 | 0.04 |
ENSRNOT00000093631
ENSRNOT00000085096 |
Rrh
|
retinal pigment epithelium derived rhodopsin homolog |
| chr7_-_2961873 | 0.04 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr13_+_71656651 | 0.04 |
ENSRNOT00000050365
|
Zfp648
|
zinc finger protein 648 |
| chr2_-_226779440 | 0.04 |
ENSRNOT00000055669
|
Fnbp1l
|
formin binding protein 1-like |
| chr5_-_136098013 | 0.04 |
ENSRNOT00000089166
|
RGD1563714
|
RGD1563714 |
| chr18_-_37986930 | 0.04 |
ENSRNOT00000025927
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr1_+_99505677 | 0.04 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr20_-_12938891 | 0.04 |
ENSRNOT00000017141
|
RGD1564149
|
similar to Protein C21orf58 |
| chr6_+_86862790 | 0.04 |
ENSRNOT00000093492
|
Fancm
|
Fanconi anemia, complementation group M |
| chr2_+_187162017 | 0.04 |
ENSRNOT00000080693
|
Insrr
|
insulin receptor-related receptor |
| chr18_-_27395603 | 0.04 |
ENSRNOT00000034385
|
LOC100359679
|
rCG49431-like |
| chr4_-_164123974 | 0.04 |
ENSRNOT00000082447
|
LOC502907
|
similar to immunoreceptor Ly49si1 |
| chr1_+_84584596 | 0.04 |
ENSRNOT00000032933
|
LOC100910046
|
zinc finger protein 60-like |
| chrX_-_79909678 | 0.04 |
ENSRNOT00000050336
|
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr10_+_11100917 | 0.04 |
ENSRNOT00000006067
|
Coro7
|
coronin 7 |
| chr6_-_111477090 | 0.04 |
ENSRNOT00000016389
|
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr5_-_159946446 | 0.04 |
ENSRNOT00000089184
|
Clcnka
|
chloride voltage-gated channel Ka |
| chr11_-_43099412 | 0.04 |
ENSRNOT00000002281
|
Gabrr3
|
gamma-aminobutyric acid type A receptor rho3 subunit |
| chr13_-_88436789 | 0.04 |
ENSRNOT00000003901
|
Ddr2
|
discoidin domain receptor tyrosine kinase 2 |
| chr3_+_92403582 | 0.04 |
ENSRNOT00000064282
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr7_+_27174882 | 0.04 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr15_-_71779033 | 0.04 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr9_+_73687743 | 0.04 |
ENSRNOT00000079495
ENSRNOT00000017197 |
Rpe
|
ribulose-5-phosphate-3-epimerase |
| chr2_+_206454208 | 0.04 |
ENSRNOT00000026863
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr20_+_13498926 | 0.04 |
ENSRNOT00000070992
ENSRNOT00000045375 |
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b |
| chr15_+_44441856 | 0.04 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr1_+_116604550 | 0.04 |
ENSRNOT00000083587
|
Ube3a
|
ubiquitin protein ligase E3A |
| chr15_-_8914501 | 0.04 |
ENSRNOT00000008752
|
Thrb
|
thyroid hormone receptor beta |
| chr10_-_31790263 | 0.04 |
ENSRNOT00000059468
|
Timd2
|
T-cell immunoglobulin and mucin domain containing 2 |
| chr7_+_13151592 | 0.04 |
ENSRNOT00000010203
|
Vom2r53
|
vomeronasal 2 receptor, 53 |
| chr3_+_11317183 | 0.04 |
ENSRNOT00000091171
ENSRNOT00000016341 |
Golga2
|
golgin A2 |
| chr17_-_43770561 | 0.04 |
ENSRNOT00000088408
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr1_+_85437780 | 0.04 |
ENSRNOT00000093740
|
Supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr13_+_79269973 | 0.04 |
ENSRNOT00000003969
|
Tnfsf4
|
tumor necrosis factor superfamily member 4 |
| chr5_-_139933764 | 0.04 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chr10_+_99437436 | 0.03 |
ENSRNOT00000006254
|
Kcnj2
|
potassium voltage-gated channel subfamily J member 2 |
| chr8_+_69127708 | 0.03 |
ENSRNOT00000013490
|
Snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr8_+_21611319 | 0.03 |
ENSRNOT00000068461
|
Zfp846
|
zinc finger protein 846 |
| chr11_-_61287914 | 0.03 |
ENSRNOT00000086286
|
Spice1
|
spindle and centriole associated protein 1 |
| chr14_-_3446611 | 0.03 |
ENSRNOT00000080901
|
Brdt
|
bromodomain testis associated |
| chr1_+_114258719 | 0.03 |
ENSRNOT00000088459
ENSRNOT00000016376 |
Cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr9_+_30419001 | 0.03 |
ENSRNOT00000018116
|
Col9a1
|
collagen type IX alpha 1 chain |
| chr8_+_93439648 | 0.03 |
ENSRNOT00000043008
|
LOC100359563
|
ribosomal protein S20-like |
| chr14_-_114649173 | 0.03 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr4_-_168517177 | 0.03 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr2_+_72006099 | 0.03 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr17_+_35396286 | 0.03 |
ENSRNOT00000089613
ENSRNOT00000092638 |
Exoc2
|
exocyst complex component 2 |
| chr20_+_9791171 | 0.03 |
ENSRNOT00000078031
|
Abcg1
|
ATP binding cassette subfamily G member 1 |
| chr2_-_59084059 | 0.03 |
ENSRNOT00000086323
ENSRNOT00000088701 |
Spef2
|
sperm flagellar 2 |
| chr10_-_55560422 | 0.03 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr2_+_118932209 | 0.03 |
ENSRNOT00000077170
|
Mfn1
|
mitofusin 1 |
| chr18_+_36829062 | 0.03 |
ENSRNOT00000025467
ENSRNOT00000086979 |
Tcerg1
|
transcription elongation regulator 1 |
| chr7_+_70946228 | 0.03 |
ENSRNOT00000039306
|
Stat6
|
signal transducer and activator of transcription 6 |
| chr9_+_65534704 | 0.03 |
ENSRNOT00000016730
|
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr14_+_108415068 | 0.03 |
ENSRNOT00000083763
|
Pus10
|
pseudouridylate synthase 10 |
| chr11_+_54404446 | 0.03 |
ENSRNOT00000002678
|
Dzip3
|
DAZ interacting zinc finger protein 3 |
| chr8_-_49502647 | 0.03 |
ENSRNOT00000040313
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr1_-_84732242 | 0.03 |
ENSRNOT00000025586
|
LOC102553760
|
zinc finger protein 59-like |
| chr18_-_27550224 | 0.03 |
ENSRNOT00000037368
|
Cdc25c
|
cell division cycle 25C |
| chr8_-_87315955 | 0.03 |
ENSRNOT00000081437
|
Filip1
|
filamin A interacting protein 1 |
| chr14_+_8182383 | 0.03 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chrX_-_139329975 | 0.03 |
ENSRNOT00000086405
|
LOC103694517
|
high mobility group protein B4-like |
| chr6_+_9790422 | 0.03 |
ENSRNOT00000020959
|
Prkce
|
protein kinase C, epsilon |
| chr2_+_211546560 | 0.03 |
ENSRNOT00000033443
|
Aknad1
|
AKNA domain containing 1 |
| chr12_-_38525432 | 0.03 |
ENSRNOT00000011240
|
Lrrc43
|
leucine rich repeat containing 43 |
| chr1_+_87563975 | 0.03 |
ENSRNOT00000088772
|
Zfp30
|
zinc finger protein 30 |
| chr9_+_46997798 | 0.03 |
ENSRNOT00000087112
ENSRNOT00000082408 |
Il1r1
|
interleukin 1 receptor type 1 |
| chr9_-_30844199 | 0.03 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr20_-_27308069 | 0.03 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr9_+_50925619 | 0.03 |
ENSRNOT00000076248
ENSRNOT00000059349 |
Ercc5
|
ERCC excision repair 5, endonuclease |
| chr5_-_161879133 | 0.03 |
ENSRNOT00000041175
|
Prdm2
|
PR/SET domain 2 |
| chr4_+_140092352 | 0.03 |
ENSRNOT00000008892
|
Setmar
|
SET domain and mariner transposase fusion gene |
| chr7_-_27213651 | 0.03 |
ENSRNOT00000077041
ENSRNOT00000076468 |
Tdg
|
thymine-DNA glycosylase |
| chr9_-_49902943 | 0.03 |
ENSRNOT00000037094
|
Tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr2_+_243422811 | 0.03 |
ENSRNOT00000014694
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr17_+_34665872 | 0.03 |
ENSRNOT00000092101
ENSRNOT00000085704 |
Exoc2
|
exocyst complex component 2 |
| chr4_-_155763500 | 0.03 |
ENSRNOT00000071745
|
LOC100909595
|
solute carrier family 2, facilitated glucose transporter member 3-like |
| chr1_+_72874583 | 0.03 |
ENSRNOT00000077719
|
Dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr6_+_111642411 | 0.03 |
ENSRNOT00000016962
|
Adck1
|
aarF domain containing kinase 1 |
| chr1_-_266074181 | 0.03 |
ENSRNOT00000026378
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr6_+_37555182 | 0.03 |
ENSRNOT00000082616
|
Fam49a
|
family with sequence similarity 49, member A |
| chr3_+_160092975 | 0.03 |
ENSRNOT00000080707
|
Pkig
|
cAMP-dependent protein kinase inhibitor gamma |
| chr6_+_33517769 | 0.03 |
ENSRNOT00000007991
|
Hs1bp3
|
HCLS1 binding protein 3 |
| chr17_+_19249952 | 0.03 |
ENSRNOT00000023140
|
Atxn1
|
ataxin 1 |
| chr6_+_47916188 | 0.03 |
ENSRNOT00000011819
|
Rnaseh1
|
ribonuclease H1 |
| chr2_+_92573004 | 0.03 |
ENSRNOT00000065774
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr9_-_17209220 | 0.03 |
ENSRNOT00000083764
|
Gtpbp2
|
GTP binding protein 2 |
| chr11_+_57207656 | 0.03 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_-_77031120 | 0.03 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chr2_+_247299433 | 0.02 |
ENSRNOT00000050330
|
Unc5c
|
unc-5 netrin receptor C |
| chrM_+_3904 | 0.02 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr6_-_50923510 | 0.02 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chrM_+_9451 | 0.02 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr1_-_84950210 | 0.02 |
ENSRNOT00000083050
|
AABR07002784.1
|
|
| chr1_-_282492912 | 0.02 |
ENSRNOT00000047883
|
AABR07007130.1
|
|
| chr10_-_35040423 | 0.02 |
ENSRNOT00000073738
|
Trappc2b
|
trafficking protein particle complex 2B |
| chr8_+_50139997 | 0.02 |
ENSRNOT00000022796
|
Bace1
|
beta-secretase 1 |
| chr4_+_88694583 | 0.02 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr9_-_94192813 | 0.02 |
ENSRNOT00000025978
|
Alpp
|
alkaline phosphatase, placental |
| chr4_-_100213858 | 0.02 |
ENSRNOT00000015562
|
Usp39
|
ubiquitin specific peptidase 39 |
| chr10_-_91807937 | 0.02 |
ENSRNOT00000005074
|
Wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr2_-_192027225 | 0.02 |
ENSRNOT00000016430
|
Prr9
|
proline rich 9 |
| chr14_-_60503803 | 0.02 |
ENSRNOT00000006093
|
Anapc4
|
anaphase promoting complex subunit 4 |
| chr7_-_15073052 | 0.02 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chrX_-_14910727 | 0.02 |
ENSRNOT00000085079
ENSRNOT00000030146 |
Ssx1
|
SSX family member 1 |
| chr20_+_2044200 | 0.02 |
ENSRNOT00000092626
|
RT1-M4
|
RT1 class Ib, locus M4 |
| chr10_+_47019326 | 0.02 |
ENSRNOT00000006984
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr10_-_25890639 | 0.02 |
ENSRNOT00000085499
|
Hmmr
|
hyaluronan-mediated motility receptor |
| chr10_-_85709568 | 0.02 |
ENSRNOT00000005426
|
Cwc25
|
CWC25 spliceosome-associated protein homolog |
| chr10_-_37311625 | 0.02 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr1_+_48077033 | 0.02 |
ENSRNOT00000020100
|
Mas1
|
MAS1 proto-oncogene, G protein-coupled receptor |
| chr1_-_100359511 | 0.02 |
ENSRNOT00000056420
|
RGD1309036
|
hypothetical LOC292874 |
| chr10_+_13145099 | 0.02 |
ENSRNOT00000091287
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr1_+_213676954 | 0.02 |
ENSRNOT00000050551
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr8_+_55050284 | 0.02 |
ENSRNOT00000013242
|
Pih1d2
|
PIH1 domain containing 2 |
| chr1_-_87147308 | 0.02 |
ENSRNOT00000027773
ENSRNOT00000089305 ENSRNOT00000090402 |
Actn4
|
actinin alpha 4 |
| chr14_+_9226125 | 0.02 |
ENSRNOT00000088047
|
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr10_-_58693754 | 0.02 |
ENSRNOT00000071764
|
Pitpnm3
|
PITPNM family member 3 |
| chr15_+_52767442 | 0.02 |
ENSRNOT00000014441
|
LOC108348161
|
phytanoyl-CoA hydroxylase-interacting protein |
| chr1_+_129255396 | 0.02 |
ENSRNOT00000040147
|
Fam169b
|
family with sequence similarity 169, member B |
| chr12_-_17322608 | 0.02 |
ENSRNOT00000033038
|
LOC102546864
|
uncharacterized LOC102546864 |
| chr10_-_62698541 | 0.02 |
ENSRNOT00000019759
|
Coro6
|
coronin 6 |
| chr2_-_227207584 | 0.02 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr1_+_265157379 | 0.02 |
ENSRNOT00000022426
|
Btrc
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chrX_+_35599258 | 0.02 |
ENSRNOT00000072627
ENSRNOT00000005061 |
Cdkl5
|
cyclin-dependent kinase-like 5 |
| chr1_+_15782477 | 0.02 |
ENSRNOT00000088025
ENSRNOT00000089966 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr1_+_256955652 | 0.02 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr8_+_118402930 | 0.02 |
ENSRNOT00000037662
|
LOC100911725
|
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4-like |
| chr1_+_277261681 | 0.02 |
ENSRNOT00000022724
|
Plekhs1
|
pleckstrin homology domain containing S1 |
| chr9_+_95221474 | 0.02 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr3_-_80003032 | 0.02 |
ENSRNOT00000017360
|
Madd
|
MAP-kinase activating death domain |
Network of associatons between targets according to the STRING database.
First level regulatory network of Obox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:1903406 | positive regulation of synaptic growth at neuromuscular junction(GO:0045887) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of protein geranylgeranylation(GO:2000539) positive regulation of protein geranylgeranylation(GO:2000541) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:2000292 | regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.0 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.0 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.0 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.0 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.0 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.0 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) corticotropin-releasing hormone receptor activity(GO:0043404) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |