Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
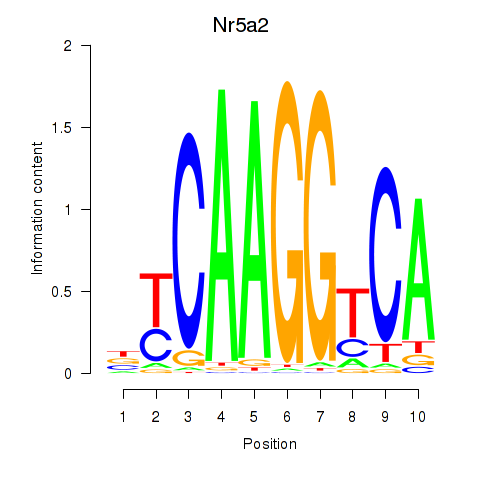
Results for Nr5a2
Z-value: 1.08
Transcription factors associated with Nr5a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr5a2
|
ENSRNOG00000000653 | nuclear receptor subfamily 5, group A, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr5a2 | rn6_v1_chr13_-_53870428_53870428 | 0.15 | 8.1e-01 | Click! |
Activity profile of Nr5a2 motif
Sorted Z-values of Nr5a2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_198360678 | 0.88 |
ENSRNOT00000051917
|
Hist2h2ac
|
histone cluster 2 H2A family member c |
| chr5_+_50381244 | 0.49 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr17_+_44520537 | 0.48 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr3_+_147609095 | 0.42 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr10_+_110139783 | 0.38 |
ENSRNOT00000054939
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr14_+_43694183 | 0.37 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chr15_+_41069507 | 0.36 |
ENSRNOT00000018533
|
C1qtnf9
|
C1q and tumor necrosis factor related protein 9 |
| chr1_+_276659542 | 0.35 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr1_+_72882806 | 0.34 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr17_-_44758170 | 0.33 |
ENSRNOT00000091176
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr15_-_33656089 | 0.33 |
ENSRNOT00000024186
|
Myh7
|
myosin heavy chain 7 |
| chr2_-_187742747 | 0.30 |
ENSRNOT00000026530
|
Bglap
|
bone gamma-carboxyglutamate protein |
| chr4_-_123118186 | 0.29 |
ENSRNOT00000038096
|
LOC100361898
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr4_+_165732643 | 0.29 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr10_-_97582188 | 0.29 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr3_+_58164931 | 0.29 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr20_-_5618254 | 0.27 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr7_+_12229379 | 0.26 |
ENSRNOT00000060811
|
Adamtsl5
|
ADAMTS-like 5 |
| chr17_-_44834305 | 0.25 |
ENSRNOT00000084303
|
Hist1h2bd
|
histone cluster 1, H2bd |
| chr1_-_167005839 | 0.24 |
ENSRNOT00000027261
|
Lrrc51
|
leucine rich repeat containing 51 |
| chr6_+_55085313 | 0.24 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chr11_+_64472072 | 0.22 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr8_-_128266639 | 0.22 |
ENSRNOT00000064555
ENSRNOT00000066932 |
Scn5a
|
sodium voltage-gated channel alpha subunit 5 |
| chr1_-_89258935 | 0.22 |
ENSRNOT00000003180
|
LOC100361079
|
ribosomal protein L36-like |
| chr12_+_47024442 | 0.21 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr1_+_221773254 | 0.21 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr10_+_64930023 | 0.20 |
ENSRNOT00000071102
|
AABR07030053.1
|
|
| chr4_+_145427367 | 0.20 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chr9_+_43259709 | 0.20 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr7_-_121311565 | 0.19 |
ENSRNOT00000092252
|
Rpl3
|
ribosomal protein L3 |
| chr5_-_151397603 | 0.19 |
ENSRNOT00000076866
|
Gpr3
|
G protein-coupled receptor 3 |
| chr17_+_44758460 | 0.19 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chr17_+_43627930 | 0.18 |
ENSRNOT00000081719
|
LOC102549061
|
histone H2B type 1-N-like |
| chr9_+_61692154 | 0.18 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr17_-_89163113 | 0.17 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr14_+_73674753 | 0.17 |
ENSRNOT00000040676
|
RGD1561694
|
similar to High mobility group protein 2 (HMG-2) |
| chr5_+_64790186 | 0.17 |
ENSRNOT00000078502
|
Zfp189
|
zinc finger protein 189 |
| chr1_-_222468896 | 0.16 |
ENSRNOT00000028754
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr11_+_70687500 | 0.15 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr5_-_136002900 | 0.15 |
ENSRNOT00000025197
|
Plk3
|
polo-like kinase 3 |
| chr8_+_118378059 | 0.15 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr19_-_57699113 | 0.15 |
ENSRNOT00000026767
|
Egln1
|
egl-9 family hypoxia-inducible factor 1 |
| chr5_+_173640780 | 0.15 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr3_-_123702732 | 0.15 |
ENSRNOT00000028859
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chrX_+_15273933 | 0.14 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr17_-_44748188 | 0.13 |
ENSRNOT00000081970
|
LOC103690190
|
histone H2A type 1-E |
| chr5_-_150323063 | 0.13 |
ENSRNOT00000014084
|
Oprd1
|
opioid receptor, delta 1 |
| chr6_-_6842758 | 0.13 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr9_-_15628881 | 0.13 |
ENSRNOT00000077318
ENSRNOT00000020950 |
Guca1b
|
guanylate cyclase activator 1B |
| chr2_-_186245771 | 0.12 |
ENSRNOT00000066004
ENSRNOT00000079954 |
Dclk2
|
doublecortin-like kinase 2 |
| chr10_+_45289741 | 0.12 |
ENSRNOT00000066190
|
Hist3h2ba
|
histone cluster 3, H2ba |
| chr2_+_206064179 | 0.12 |
ENSRNOT00000025953
|
Syt6
|
synaptotagmin 6 |
| chr7_+_129860327 | 0.12 |
ENSRNOT00000043461
|
Pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr5_+_64326733 | 0.12 |
ENSRNOT00000065775
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chrX_-_139916883 | 0.12 |
ENSRNOT00000090442
|
Gpc3
|
glypican 3 |
| chr8_-_118378460 | 0.11 |
ENSRNOT00000047247
|
RGD1563705
|
similar to ribosomal protein S23 |
| chr14_-_84937725 | 0.11 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr4_-_148845267 | 0.11 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr19_-_10653800 | 0.11 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr7_-_119158173 | 0.11 |
ENSRNOT00000067483
ENSRNOT00000078528 |
Txn2
|
thioredoxin 2 |
| chr11_+_37798370 | 0.11 |
ENSRNOT00000002679
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr13_-_85622314 | 0.11 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr16_-_7681576 | 0.10 |
ENSRNOT00000026550
|
Colq
|
collagen like tail subunit of asymmetric acetylcholinesterase |
| chr2_-_119140110 | 0.10 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
| chr7_+_121889157 | 0.10 |
ENSRNOT00000024835
|
Fam83f
|
family with sequence similarity 83, member F |
| chr5_+_157423213 | 0.10 |
ENSRNOT00000023431
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr1_+_197813963 | 0.10 |
ENSRNOT00000045531
|
LOC100361060
|
ribosomal protein L36-like |
| chr16_+_19874034 | 0.09 |
ENSRNOT00000023579
|
Mrpl34
|
mitochondrial ribosomal protein L34 |
| chr10_+_109658032 | 0.09 |
ENSRNOT00000054965
|
Mrpl12
|
mitochondrial ribosomal protein L12 |
| chr19_-_37427989 | 0.09 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr6_+_126434226 | 0.09 |
ENSRNOT00000090857
|
Chga
|
chromogranin A |
| chr7_+_35125424 | 0.09 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr2_-_198706428 | 0.09 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr8_+_117280705 | 0.09 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr5_-_157573183 | 0.08 |
ENSRNOT00000064418
|
Minos1
|
mitochondrial inner membrane organizing system 1 |
| chr4_+_67378188 | 0.08 |
ENSRNOT00000030892
|
Ndufb2
|
NADH:ubiquinone oxidoreductase subunit B2 |
| chr7_-_12424367 | 0.08 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr19_-_54245855 | 0.08 |
ENSRNOT00000023855
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr1_-_173764246 | 0.08 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr8_-_130491998 | 0.08 |
ENSRNOT00000064114
|
Higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr12_-_2545740 | 0.08 |
ENSRNOT00000075527
|
Tgfbr3l
|
transforming growth factor beta receptor 3 like |
| chr19_+_26084903 | 0.08 |
ENSRNOT00000004799
|
Prdx2
|
peroxiredoxin 2 |
| chr14_+_89314176 | 0.08 |
ENSRNOT00000006765
|
Upp1
|
uridine phosphorylase 1 |
| chr7_+_2504695 | 0.08 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr1_-_218374619 | 0.08 |
ENSRNOT00000017860
|
Rmt1
|
mammary cancer associated protein RMT-1 |
| chr3_-_79728879 | 0.07 |
ENSRNOT00000012425
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr6_-_135412312 | 0.07 |
ENSRNOT00000010610
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr20_-_48294948 | 0.07 |
ENSRNOT00000072716
|
LOC683897
|
similar to Protein C6orf203 |
| chr9_+_10471742 | 0.07 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr8_+_116094851 | 0.07 |
ENSRNOT00000084120
|
RGD1307461
|
similar to RIKEN cDNA 6430571L13 gene; similar to g20 protein |
| chr5_-_160403373 | 0.07 |
ENSRNOT00000018393
|
Ctrc
|
chymotrypsin C |
| chr10_+_67862054 | 0.07 |
ENSRNOT00000031746
|
Cdk5r1
|
cyclin-dependent kinase 5 regulatory subunit 1 |
| chr18_+_83777665 | 0.07 |
ENSRNOT00000018682
|
Cbln2
|
cerebellin 2 precursor |
| chr7_-_11330278 | 0.07 |
ENSRNOT00000027730
|
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr5_+_150032999 | 0.07 |
ENSRNOT00000013301
|
Srsf4
|
serine and arginine rich splicing factor 4 |
| chr2_-_140464607 | 0.07 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr13_+_52413241 | 0.07 |
ENSRNOT00000035157
|
AABR07020999.1
|
|
| chr9_-_10471009 | 0.07 |
ENSRNOT00000072868
|
Safb
|
scaffold attachment factor B |
| chr1_-_143535583 | 0.07 |
ENSRNOT00000087785
|
Homer2
|
homer scaffolding protein 2 |
| chr3_+_94549180 | 0.06 |
ENSRNOT00000016318
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr9_+_17705212 | 0.06 |
ENSRNOT00000026728
|
Tmem63b
|
transmembrane protein 63B |
| chr8_-_119157071 | 0.06 |
ENSRNOT00000028963
|
Tmie
|
transmembrane inner ear |
| chr3_+_93495106 | 0.06 |
ENSRNOT00000029922
ENSRNOT00000085760 |
Abtb2
|
ankyrin repeat and BTB domain containing 2 |
| chrX_+_68774699 | 0.06 |
ENSRNOT00000081662
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr13_-_36101411 | 0.06 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr6_-_136145837 | 0.06 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr15_-_36114752 | 0.06 |
ENSRNOT00000045209
|
Gzmbl3
|
Granzyme B-like 3 |
| chr2_-_183031214 | 0.06 |
ENSRNOT00000013260
|
LOC679811
|
similar to RIKEN cDNA D930015E06 |
| chr12_+_21767606 | 0.06 |
ENSRNOT00000059602
|
LOC103691013
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr12_+_25264192 | 0.06 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr7_+_110031696 | 0.06 |
ENSRNOT00000012753
|
Khdrbs3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr1_-_222250980 | 0.06 |
ENSRNOT00000028734
|
Nudt22
|
nudix hydrolase 22 |
| chr11_-_32088002 | 0.06 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr3_+_4083864 | 0.05 |
ENSRNOT00000006186
|
Fam69b
|
family with sequence similarity 69, member B |
| chr13_-_47831110 | 0.05 |
ENSRNOT00000061070
|
Mapkapk2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr10_+_78168715 | 0.05 |
ENSRNOT00000072294
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr2_+_154556029 | 0.05 |
ENSRNOT00000084238
|
Gmps
|
guanine monophosphate synthase |
| chr1_-_222350173 | 0.05 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr9_+_99998275 | 0.05 |
ENSRNOT00000074395
|
Gpc1
|
glypican 1 |
| chr1_+_220325352 | 0.05 |
ENSRNOT00000027258
|
LOC108348098
|
breast cancer metastasis-suppressor 1 homolog |
| chr8_-_122402127 | 0.05 |
ENSRNOT00000074238
ENSRNOT00000076548 |
Crtapl1
|
cartilage associated protein-like 1 |
| chr18_+_2416871 | 0.05 |
ENSRNOT00000081399
|
Gata6
|
GATA binding protein 6 |
| chr4_-_78879294 | 0.05 |
ENSRNOT00000084543
|
Igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr1_-_266074181 | 0.05 |
ENSRNOT00000026378
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr15_+_2631529 | 0.05 |
ENSRNOT00000018897
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr18_-_27749235 | 0.05 |
ENSRNOT00000026696
|
Hspa9
|
heat shock protein family A member 9 |
| chr19_-_11326139 | 0.05 |
ENSRNOT00000025669
|
Mt3
|
metallothionein 3 |
| chr2_+_119139717 | 0.04 |
ENSRNOT00000016051
|
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr4_+_66405166 | 0.04 |
ENSRNOT00000077486
ENSRNOT00000067097 |
Clec2l
|
C-type lectin domain family 2, member L |
| chr5_-_173209768 | 0.04 |
ENSRNOT00000045053
|
Atad3a
|
ATPase family, AAA domain containing 3A |
| chr3_+_11607225 | 0.04 |
ENSRNOT00000072685
|
St6galnac4
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr2_-_88113029 | 0.04 |
ENSRNOT00000013354
|
Car2
|
carbonic anhydrase 2 |
| chr5_-_155772040 | 0.04 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr2_+_154555871 | 0.04 |
ENSRNOT00000076292
|
Gmps
|
guanine monophosphate synthase |
| chr14_+_84953786 | 0.04 |
ENSRNOT00000010332
|
Zmat5
|
zinc finger, matrin type 5 |
| chr3_+_113319456 | 0.04 |
ENSRNOT00000051354
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr1_+_252375933 | 0.04 |
ENSRNOT00000078405
|
Lipn
|
lipase, family member N |
| chr3_+_113318563 | 0.04 |
ENSRNOT00000089230
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr9_+_81566074 | 0.04 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr19_+_52521809 | 0.04 |
ENSRNOT00000081019
|
Klhl36
|
kelch-like family member 36 |
| chr6_+_128750795 | 0.04 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr3_+_133232432 | 0.04 |
ENSRNOT00000006412
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr7_+_130308532 | 0.04 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr14_-_43694584 | 0.04 |
ENSRNOT00000041866
|
AABR07072400.1
|
|
| chr9_+_40975836 | 0.04 |
ENSRNOT00000084470
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr19_+_54245950 | 0.04 |
ENSRNOT00000024033
|
Cox4i1
|
cytochrome c oxidase subunit 4i1 |
| chr14_+_86673775 | 0.04 |
ENSRNOT00000091873
ENSRNOT00000079015 |
Ppia
|
peptidylprolyl isomerase A |
| chr7_-_120518653 | 0.04 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr7_+_11724962 | 0.03 |
ENSRNOT00000026551
|
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_-_175586802 | 0.03 |
ENSRNOT00000071333
|
AABR07005027.1
|
|
| chrX_-_15707436 | 0.03 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr15_-_52134140 | 0.03 |
ENSRNOT00000013390
|
Polr3d
|
RNA polymerase III subunit D |
| chr7_+_121311024 | 0.03 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr17_+_49714294 | 0.03 |
ENSRNOT00000018190
|
Rala
|
RAS like proto-oncogene A |
| chr5_+_159484370 | 0.03 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr1_-_72674271 | 0.03 |
ENSRNOT00000023744
|
Kmt5c
|
lysine methyltransferase 5C |
| chr17_-_43627629 | 0.03 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr1_-_134870255 | 0.03 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chrX_+_105575759 | 0.03 |
ENSRNOT00000088760
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr9_+_17911944 | 0.03 |
ENSRNOT00000080940
|
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr10_-_15235740 | 0.03 |
ENSRNOT00000027170
|
Mcrip2
|
MAPK regulated co-repressor interacting protein 2 |
| chr11_+_86890585 | 0.03 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr4_-_157679962 | 0.03 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_142136452 | 0.03 |
ENSRNOT00000016445
|
Hddc3
|
HD domain containing 3 |
| chr1_-_220467159 | 0.03 |
ENSRNOT00000075365
|
Tmem151a
|
transmembrane protein 151A |
| chr1_+_224882439 | 0.02 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr4_+_7260575 | 0.02 |
ENSRNOT00000064893
|
Fastk
|
Fas-activated serine/threonine kinase |
| chr4_-_176720012 | 0.02 |
ENSRNOT00000017965
|
Ldhb
|
lactate dehydrogenase B |
| chr16_+_16949232 | 0.02 |
ENSRNOT00000047499
|
AABR07024795.1
|
|
| chr10_+_55675575 | 0.02 |
ENSRNOT00000057295
|
Vamp2
|
vesicle-associated membrane protein 2 |
| chr7_-_117576737 | 0.02 |
ENSRNOT00000039795
|
Dgat1
|
diacylglycerol O-acyltransferase 1 |
| chr10_+_39066716 | 0.02 |
ENSRNOT00000010729
|
Il5
|
interleukin 5 |
| chr6_+_58388333 | 0.02 |
ENSRNOT00000036337
|
LOC680646
|
similar to 40S ribosomal protein S2 |
| chr19_-_21492468 | 0.02 |
ENSRNOT00000087660
ENSRNOT00000048345 ENSRNOT00000020770 |
Lonp2
|
lon peptidase 2, peroxisomal |
| chr8_-_76239995 | 0.02 |
ENSRNOT00000071321
|
LOC100912027
|
60S ribosomal protein L27-like |
| chr9_-_46309451 | 0.02 |
ENSRNOT00000018684
|
Rnf149
|
ring finger protein 149 |
| chr5_-_60658521 | 0.02 |
ENSRNOT00000092866
ENSRNOT00000093024 ENSRNOT00000016875 |
Tomm5
Fbxo10
|
translocase of outer mitochondrial membrane 5 F-box protein 10 |
| chr3_-_141411170 | 0.02 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr11_+_61661310 | 0.02 |
ENSRNOT00000093325
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr11_-_72109964 | 0.01 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr7_-_74735650 | 0.01 |
ENSRNOT00000014407
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr15_-_5894854 | 0.01 |
ENSRNOT00000024427
|
Cd99l2
|
CD99 molecule-like 2 |
| chr4_+_9821541 | 0.01 |
ENSRNOT00000015733
|
Slc26a5
|
solute carrier family 26 member 5 |
| chr12_-_30566032 | 0.01 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr14_-_85318194 | 0.01 |
ENSRNOT00000065135
|
Gas2l1
|
growth arrest-specific 2 like 1 |
| chr20_-_29029905 | 0.01 |
ENSRNOT00000075682
|
Oit3
|
oncoprotein induced transcript 3 |
| chr13_-_51076852 | 0.01 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr5_+_140923914 | 0.01 |
ENSRNOT00000020929
|
Heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr15_-_35417273 | 0.01 |
ENSRNOT00000083961
ENSRNOT00000041430 |
Gzmb
|
granzyme B |
| chr1_-_165606375 | 0.01 |
ENSRNOT00000024290
|
Mrpl48
|
mitochondrial ribosomal protein L48 |
| chr19_+_22569999 | 0.01 |
ENSRNOT00000022573
|
Dnaja2
|
DnaJ heat shock protein family (Hsp40) member A2 |
| chr4_-_80333326 | 0.01 |
ENSRNOT00000014058
|
LOC100363502
|
cytochrome c, somatic-like |
| chr3_-_3476215 | 0.01 |
ENSRNOT00000024352
|
Ubac1
|
UBA domain containing 1 |
| chr15_+_5265307 | 0.01 |
ENSRNOT00000031807
|
LOC108348081
|
CD99 antigen-like protein 2 |
| chr2_-_210873024 | 0.01 |
ENSRNOT00000026051
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr20_+_21316826 | 0.01 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chr4_+_145653602 | 0.01 |
ENSRNOT00000038964
|
Tatdn2
|
TatD DNase domain containing 2 |
| chr8_-_116469915 | 0.01 |
ENSRNOT00000024193
|
Sema3f
|
semaphorin 3F |
| chr18_-_56331991 | 0.01 |
ENSRNOT00000085841
ENSRNOT00000091220 |
Slc6a7
|
solute carrier family 6 member 7 |
| chr9_-_100888107 | 0.01 |
ENSRNOT00000024794
|
Thap4
|
THAP domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr5a2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048619 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) response to 3,3',5-triiodo-L-thyronine(GO:1905242) cellular response to 3,3',5-triiodo-L-thyronine(GO:1905243) |
| 0.1 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.2 | GO:2000777 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.3 | GO:0048597 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.4 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0098912 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.3 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.0 | 0.5 | GO:0046884 | thyroid hormone generation(GO:0006590) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0061209 | cell proliferation involved in mesonephros development(GO:0061209) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 1.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.3 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 0.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.1 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 | 0.0 | GO:2000295 | cadmium ion homeostasis(GO:0055073) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.0 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0003922 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |