Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Nr4a3
Z-value: 0.21
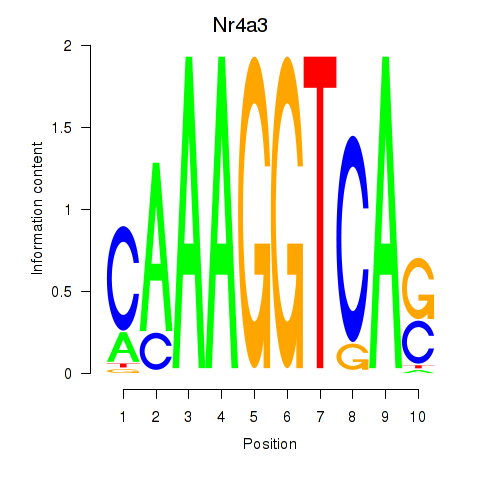
Transcription factors associated with Nr4a3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a3
|
ENSRNOG00000005964 | nuclear receptor subfamily 4, group A, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a3 | rn6_v1_chr5_+_63781801_63781801 | 0.91 | 3.3e-02 | Click! |
Activity profile of Nr4a3 motif
Sorted Z-values of Nr4a3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_55891710 | 0.08 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr11_+_83868655 | 0.07 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr6_+_135610743 | 0.07 |
ENSRNOT00000010906
|
Traf3
|
Tnf receptor-associated factor 3 |
| chr2_-_207300854 | 0.07 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr1_-_141579871 | 0.05 |
ENSRNOT00000020002
|
Anpep
|
alanyl aminopeptidase, membrane |
| chr3_-_72113680 | 0.05 |
ENSRNOT00000009708
|
Zdhhc5
|
zinc finger, DHHC-type containing 5 |
| chr15_-_14622587 | 0.04 |
ENSRNOT00000086502
|
Synpr
|
synaptoporin |
| chr1_-_141188031 | 0.04 |
ENSRNOT00000044567
|
Polg
|
DNA polymerase gamma, catalytic subunit |
| chr1_-_259484569 | 0.04 |
ENSRNOT00000021289
ENSRNOT00000021114 ENSRNOT00000021229 ENSRNOT00000021347 ENSRNOT00000021226 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr8_+_116804451 | 0.04 |
ENSRNOT00000041402
|
Ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr20_+_3351303 | 0.04 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr20_+_3827367 | 0.03 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr1_+_196996581 | 0.03 |
ENSRNOT00000021690
|
Il21r
|
interleukin 21 receptor |
| chr1_-_16203838 | 0.03 |
ENSRNOT00000018556
ENSRNOT00000078586 |
Pde7b
|
phosphodiesterase 7B |
| chr4_-_15505362 | 0.03 |
ENSRNOT00000009763
|
Hgf
|
hepatocyte growth factor |
| chr10_-_49009156 | 0.03 |
ENSRNOT00000072970
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr15_-_106678918 | 0.02 |
ENSRNOT00000034723
|
Stk24
|
serine/threonine kinase 24 |
| chr20_+_3827086 | 0.02 |
ENSRNOT00000081588
|
Rxrb
|
retinoid X receptor beta |
| chr1_+_226091774 | 0.02 |
ENSRNOT00000027693
|
Fads3
|
fatty acid desaturase 3 |
| chr2_+_196304492 | 0.02 |
ENSRNOT00000028640
|
Lysmd1
|
LysM domain containing 1 |
| chr1_-_219544328 | 0.01 |
ENSRNOT00000025847
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chrX_-_4945944 | 0.01 |
ENSRNOT00000077238
|
Kdm6a
|
lysine demethylase 6A |
| chr1_-_166988062 | 0.01 |
ENSRNOT00000039564
|
Lrtomt
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr4_+_64088900 | 0.01 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr3_-_81304181 | 0.01 |
ENSRNOT00000079746
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr1_+_265904566 | 0.01 |
ENSRNOT00000086041
ENSRNOT00000036156 |
Gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr7_-_117259791 | 0.01 |
ENSRNOT00000086550
|
Plec
|
plectin |
| chr12_-_46718355 | 0.01 |
ENSRNOT00000030031
|
Rab35
|
RAB35, member RAS oncogene family |
| chr4_+_172942020 | 0.01 |
ENSRNOT00000072450
|
Lmo3
|
LIM domain only 3 |
| chr4_-_131694755 | 0.01 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr3_-_92749121 | 0.01 |
ENSRNOT00000008760
|
Cd44
|
CD44 molecule (Indian blood group) |
| chr8_-_50531423 | 0.01 |
ENSRNOT00000090985
ENSRNOT00000074942 |
Apoc3
|
apolipoprotein C3 |
| chr9_+_64745051 | 0.01 |
ENSRNOT00000021527
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr7_-_70661891 | 0.01 |
ENSRNOT00000010240
|
Inhbc
|
inhibin beta C subunit |
| chr8_-_48449311 | 0.01 |
ENSRNOT00000010134
|
Rnf26
|
ring finger protein 26 |
| chrX_-_139916883 | 0.00 |
ENSRNOT00000090442
|
Gpc3
|
glypican 3 |
| chr19_+_57556694 | 0.00 |
ENSRNOT00000032359
|
Trim67
|
tripartite motif-containing 67 |
| chr8_-_119265157 | 0.00 |
ENSRNOT00000056100
|
Rtp3
|
receptor (chemosensory) transporter protein 3 |
| chr1_+_107262659 | 0.00 |
ENSRNOT00000022499
|
Gas2
|
growth arrest-specific 2 |
| chr17_-_80807181 | 0.00 |
ENSRNOT00000040052
ENSRNOT00000090064 |
Cubn
|
cubilin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr4a3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.0 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0052724 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |