Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
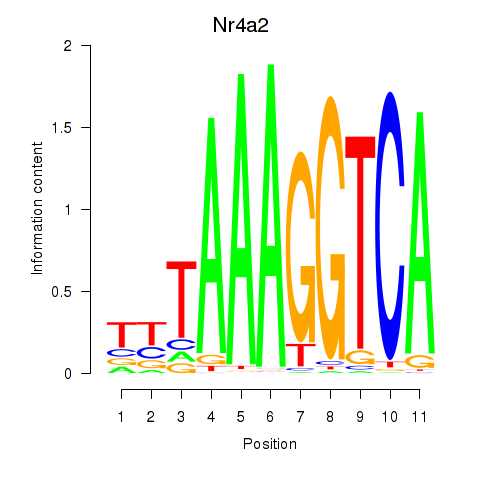
Results for Nr4a2
Z-value: 0.37
Transcription factors associated with Nr4a2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr4a2
|
ENSRNOG00000005600 | nuclear receptor subfamily 4, group A, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a2 | rn6_v1_chr3_-_43117337_43117337 | 0.50 | 3.9e-01 | Click! |
Activity profile of Nr4a2 motif
Sorted Z-values of Nr4a2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_61531416 | 0.15 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr8_+_114867062 | 0.14 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr3_+_62648447 | 0.11 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr4_-_89281222 | 0.10 |
ENSRNOT00000010898
|
Fam13a
|
family with sequence similarity 13, member A |
| chr8_+_71591360 | 0.09 |
ENSRNOT00000090096
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr1_+_219000844 | 0.09 |
ENSRNOT00000022486
|
Kmt5b
|
lysine methyltransferase 5B |
| chr3_+_131351587 | 0.09 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr3_+_80676820 | 0.08 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr3_-_60795951 | 0.07 |
ENSRNOT00000002174
|
Atf2
|
activating transcription factor 2 |
| chr7_+_35069814 | 0.07 |
ENSRNOT00000089228
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr10_-_36372436 | 0.07 |
ENSRNOT00000039827
|
Znf454
|
zinc finger protein 454 |
| chr18_+_29490981 | 0.07 |
ENSRNOT00000032316
|
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr3_-_72113680 | 0.07 |
ENSRNOT00000009708
|
Zdhhc5
|
zinc finger, DHHC-type containing 5 |
| chrX_+_22302485 | 0.07 |
ENSRNOT00000082902
|
Kdm5c
|
lysine demethylase 5C |
| chr6_-_27512287 | 0.06 |
ENSRNOT00000085937
ENSRNOT00000081278 ENSRNOT00000081323 |
Selenoi
|
selenoprotein I |
| chr7_-_40316532 | 0.06 |
ENSRNOT00000083347
|
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chr12_-_24537313 | 0.06 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chrX_+_22302703 | 0.06 |
ENSRNOT00000078617
ENSRNOT00000086952 |
Kdm5c
|
lysine demethylase 5C |
| chr1_-_71173216 | 0.06 |
ENSRNOT00000020523
|
Zfp28
|
zinc finger protein 28 |
| chr6_+_26537707 | 0.06 |
ENSRNOT00000050102
|
Zfp513
|
zinc finger protein 513 |
| chr3_+_121596791 | 0.06 |
ENSRNOT00000024694
|
Ttl
|
tubulin tyrosine ligase |
| chr7_-_81592206 | 0.06 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr3_+_155297566 | 0.06 |
ENSRNOT00000021435
ENSRNOT00000084866 |
Dhx35
|
DEAH-box helicase 35 |
| chr15_+_3996048 | 0.06 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr18_+_45023932 | 0.05 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr2_-_178389608 | 0.05 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr12_-_5822874 | 0.05 |
ENSRNOT00000075920
|
Fry
|
FRY microtubule binding protein |
| chr14_+_42714315 | 0.05 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr18_-_26211445 | 0.05 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr7_+_53630621 | 0.05 |
ENSRNOT00000067011
ENSRNOT00000080598 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr3_+_47453821 | 0.05 |
ENSRNOT00000081682
|
Tank
|
TRAF family member-associated NFKB activator |
| chr10_-_18443934 | 0.05 |
ENSRNOT00000059895
ENSRNOT00000080021 |
Ranbp17
|
RAN binding protein 17 |
| chr1_-_213973163 | 0.04 |
ENSRNOT00000024867
|
LOC100911402
|
cell cycle exit and neuronal differentiation protein 1-like |
| chr7_+_35070284 | 0.04 |
ENSRNOT00000009578
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr18_+_79813759 | 0.03 |
ENSRNOT00000021768
|
Zfp516
|
zinc finger protein 516 |
| chr1_-_81144024 | 0.03 |
ENSRNOT00000088431
|
Zfp94
|
zinc finger protein 94 |
| chr3_-_2123820 | 0.03 |
ENSRNOT00000086843
|
Ehmt1
|
euchromatic histone lysine methyltransferase 1 |
| chr7_+_117001084 | 0.03 |
ENSRNOT00000066233
|
Zfp707l1
|
zinc finger protein 707-like 1 |
| chr10_-_82399484 | 0.03 |
ENSRNOT00000082972
ENSRNOT00000055582 |
Xylt2
|
xylosyltransferase 2 |
| chr1_+_255185629 | 0.03 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr20_+_3827086 | 0.03 |
ENSRNOT00000081588
|
Rxrb
|
retinoid X receptor beta |
| chr11_-_69201380 | 0.03 |
ENSRNOT00000085618
|
Mylk
|
myosin light chain kinase |
| chr13_+_100817359 | 0.02 |
ENSRNOT00000004330
|
Tp53bp2
|
tumor protein p53 binding protein, 2 |
| chr11_+_61531571 | 0.02 |
ENSRNOT00000093467
ENSRNOT00000002727 |
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr7_+_12840938 | 0.02 |
ENSRNOT00000039232
|
Polrmt
|
RNA polymerase mitochondrial |
| chr9_-_69953182 | 0.02 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr3_+_113257688 | 0.02 |
ENSRNOT00000019320
|
Map1a
|
microtubule-associated protein 1A |
| chr19_+_49637016 | 0.02 |
ENSRNOT00000016880
|
Bco1
|
beta-carotene oxygenase 1 |
| chr7_-_117288018 | 0.02 |
ENSRNOT00000091285
|
Plec
|
plectin |
| chr9_+_117795132 | 0.02 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chrX_-_135342996 | 0.02 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr15_+_11298478 | 0.02 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr10_-_49009156 | 0.02 |
ENSRNOT00000072970
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr2_+_153830586 | 0.02 |
ENSRNOT00000042576
|
Mme
|
membrane metallo-endopeptidase |
| chr20_+_3827367 | 0.02 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr18_+_59096643 | 0.02 |
ENSRNOT00000025325
|
Wdr7
|
WD repeat domain 7 |
| chr2_-_184289126 | 0.02 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr4_-_56438142 | 0.01 |
ENSRNOT00000077079
|
Rbm28
|
RNA binding motif protein 28 |
| chr13_-_70174565 | 0.01 |
ENSRNOT00000067135
|
Rgl1
|
ral guanine nucleotide dissociation stimulator,-like 1 |
| chr3_-_159775643 | 0.01 |
ENSRNOT00000010939
|
Jph2
|
junctophilin 2 |
| chr4_-_56438465 | 0.01 |
ENSRNOT00000064007
|
Rbm28
|
RNA binding motif protein 28 |
| chr4_+_145653602 | 0.01 |
ENSRNOT00000038964
|
Tatdn2
|
TatD DNase domain containing 2 |
| chr9_+_94702129 | 0.01 |
ENSRNOT00000080930
|
Neu2
|
neuraminidase 2 |
| chr10_+_36590325 | 0.01 |
ENSRNOT00000085203
|
Zfp354a
|
zinc finger protein 354A |
| chr1_+_167373678 | 0.01 |
ENSRNOT00000027685
ENSRNOT00000088370 |
Stim1
|
stromal interaction molecule 1 |
| chr2_+_196304492 | 0.01 |
ENSRNOT00000028640
|
Lysmd1
|
LysM domain containing 1 |
| chr9_+_2202511 | 0.00 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr14_+_83889089 | 0.00 |
ENSRNOT00000078980
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr19_+_51317425 | 0.00 |
ENSRNOT00000019298
|
Cdh13
|
cadherin 13 |
| chr16_-_22561496 | 0.00 |
ENSRNOT00000016543
|
Lpl
|
lipoprotein lipase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr4a2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.0 | GO:0048880 | medulla oblongata development(GO:0021550) sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |