Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
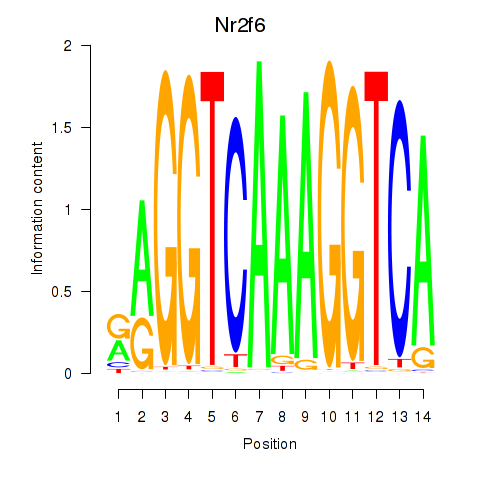
Results for Nr2f6
Z-value: 0.35
Transcription factors associated with Nr2f6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f6
|
ENSRNOG00000016892 | nuclear receptor subfamily 2, group F, member 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f6 | rn6_v1_chr16_-_19777414_19777414 | -0.44 | 4.6e-01 | Click! |
Activity profile of Nr2f6 motif
Sorted Z-values of Nr2f6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_43001948 | 0.15 |
ENSRNOT00000007374
|
Hpcal1
|
hippocalcin-like 1 |
| chr6_+_123361864 | 0.14 |
ENSRNOT00000057577
|
AABR07065353.1
|
|
| chr20_+_13827660 | 0.13 |
ENSRNOT00000093131
|
Ddt
|
D-dopachrome tautomerase |
| chr10_+_11146359 | 0.13 |
ENSRNOT00000006314
|
Pam16
|
presequence translocase associated motor 16 homolog |
| chr5_-_156734541 | 0.11 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr18_-_410098 | 0.10 |
ENSRNOT00000084138
|
LOC102546764
|
cx9C motif-containing protein 4-like |
| chr1_-_219144610 | 0.09 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr4_+_150199627 | 0.08 |
ENSRNOT00000052259
|
AABR07061871.1
|
|
| chr10_+_3227160 | 0.08 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr3_-_2938883 | 0.08 |
ENSRNOT00000084196
|
LOC108348105
|
allergen Fel d 4-like |
| chr6_+_26051396 | 0.08 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
| chr1_-_89488223 | 0.07 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr1_-_64175099 | 0.07 |
ENSRNOT00000091288
|
Ndufa3
|
NADH:ubiquinone oxidoreductase subunit A3 |
| chr7_+_117951154 | 0.07 |
ENSRNOT00000060181
|
Znf7
|
zinc finger protein 7 |
| chr15_-_45376476 | 0.07 |
ENSRNOT00000065838
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr13_-_44345735 | 0.07 |
ENSRNOT00000005006
|
Tmem163
|
transmembrane protein 163 |
| chr5_+_137257287 | 0.07 |
ENSRNOT00000037160
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr3_+_2462466 | 0.06 |
ENSRNOT00000014087
|
Rnf208
|
ring finger protein 208 |
| chr9_+_69953440 | 0.06 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr20_-_3819200 | 0.06 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr20_+_13827132 | 0.06 |
ENSRNOT00000001664
|
Ddt
|
D-dopachrome tautomerase |
| chr16_+_74810938 | 0.06 |
ENSRNOT00000058091
|
Nek5
|
NIMA-related kinase 5 |
| chr2_+_95045034 | 0.06 |
ENSRNOT00000081305
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr12_-_18526250 | 0.06 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr15_+_38069320 | 0.06 |
ENSRNOT00000014397
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr5_+_129257429 | 0.06 |
ENSRNOT00000072396
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr2_-_196304169 | 0.05 |
ENSRNOT00000028639
|
Scnm1
|
sodium channel modifier 1 |
| chr20_-_5123073 | 0.05 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr1_-_281101438 | 0.05 |
ENSRNOT00000012734
|
Rab11fip2
|
RAB11 family interacting protein 2 |
| chr11_-_61530567 | 0.05 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr2_+_239415046 | 0.05 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr10_+_89352835 | 0.05 |
ENSRNOT00000028060
|
Rpl27
|
ribosomal protein L27 |
| chr17_+_42302540 | 0.05 |
ENSRNOT00000025389
|
Gmnn
|
geminin, DNA replication inhibitor |
| chr17_+_85356042 | 0.05 |
ENSRNOT00000022201
|
Commd3
|
COMM domain containing 3 |
| chr9_-_27511176 | 0.05 |
ENSRNOT00000036813
|
LOC501110
|
similar to Glutathione S-transferase A1 (GTH1) (HA subunit 1) (GST-epsilon) (GSTA1-1) (GST class-alpha) |
| chr14_+_46001849 | 0.05 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr15_-_45947030 | 0.05 |
ENSRNOT00000035888
|
Ints6
|
integrator complex subunit 6 |
| chr1_+_189665976 | 0.05 |
ENSRNOT00000033673
|
Lyrm1
|
LYR motif containing 1 |
| chr13_-_90589466 | 0.05 |
ENSRNOT00000070812
|
Pea15
|
phosphoprotein enriched in astrocytes 15 |
| chr3_+_151691515 | 0.05 |
ENSRNOT00000052236
|
AC118414.1
|
|
| chr17_+_5281727 | 0.05 |
ENSRNOT00000024781
|
Isca1
|
iron-sulfur cluster assembly 1 |
| chr1_-_144601327 | 0.05 |
ENSRNOT00000029244
ENSRNOT00000078144 |
Saxo2
|
stabilizer of axonemal microtubules 2 |
| chr3_-_3691972 | 0.05 |
ENSRNOT00000061735
|
Qsox2
|
quiescin sulfhydryl oxidase 2 |
| chr3_-_161322289 | 0.05 |
ENSRNOT00000022594
|
Pltp
|
phospholipid transfer protein |
| chr7_-_117688187 | 0.04 |
ENSRNOT00000019707
|
Vps28
|
VPS28, ESCRT-I subunit |
| chr1_-_100231460 | 0.04 |
ENSRNOT00000032200
|
Acpt
|
acid phosphatase, testicular |
| chr1_-_226049929 | 0.04 |
ENSRNOT00000007320
|
Best1
|
bestrophin 1 |
| chr1_-_262013619 | 0.04 |
ENSRNOT00000021278
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr2_-_187706300 | 0.04 |
ENSRNOT00000092349
ENSRNOT00000026414 |
Tmem79
|
transmembrane protein 79 |
| chr6_+_76174452 | 0.04 |
ENSRNOT00000084062
|
Psma6
|
proteasome subunit alpha 6 |
| chr5_+_137257637 | 0.04 |
ENSRNOT00000093001
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr9_+_64095978 | 0.04 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr13_-_89745835 | 0.04 |
ENSRNOT00000005352
|
Klhdc9
|
kelch domain containing 9 |
| chr14_-_79464770 | 0.04 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr18_-_29504975 | 0.04 |
ENSRNOT00000063841
ENSRNOT00000024313 |
Apbb3
|
amyloid beta precursor protein binding family B member 3 |
| chr3_-_111037166 | 0.04 |
ENSRNOT00000017070
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr7_-_136853957 | 0.04 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr18_+_45023932 | 0.04 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr10_+_34975708 | 0.04 |
ENSRNOT00000064984
|
Nhp2
|
NHP2 ribonucleoprotein |
| chr14_-_86047162 | 0.03 |
ENSRNOT00000018227
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr1_+_100224401 | 0.03 |
ENSRNOT00000025846
|
Fv1
|
Friend virus susceptibility 1 |
| chr20_+_9643612 | 0.03 |
ENSRNOT00000050744
|
Umodl1
|
uromodulin-like 1 |
| chr10_+_89635675 | 0.03 |
ENSRNOT00000028179
|
RGD1561590
|
similar to SAP18 |
| chr9_-_64096265 | 0.03 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr3_+_8832740 | 0.03 |
ENSRNOT00000022552
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr11_+_86890585 | 0.03 |
ENSRNOT00000002579
|
Ranbp1
|
RAN binding protein 1 |
| chr6_-_26051229 | 0.03 |
ENSRNOT00000005855
|
Bre
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr15_-_43733182 | 0.03 |
ENSRNOT00000015318
|
Ppp2r2a
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr3_-_111037425 | 0.03 |
ENSRNOT00000085628
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr3_+_112228720 | 0.03 |
ENSRNOT00000079079
|
Capn3
|
calpain 3 |
| chr1_+_47942800 | 0.03 |
ENSRNOT00000079312
|
Wtap
|
Wilms tumor 1 associated protein |
| chr18_-_52017734 | 0.03 |
ENSRNOT00000081020
|
March3
|
membrane associated ring-CH-type finger 3 |
| chr20_+_4530342 | 0.03 |
ENSRNOT00000076352
ENSRNOT00000000478 ENSRNOT00000075925 |
Nelfe
|
negative elongation factor complex member E |
| chr11_-_70211701 | 0.03 |
ENSRNOT00000076114
|
Muc13
|
mucin 13, cell surface associated |
| chr10_-_88143965 | 0.03 |
ENSRNOT00000080167
|
Ka11
|
type I keratin KA11 |
| chr4_+_157230769 | 0.03 |
ENSRNOT00000091464
ENSRNOT00000017472 |
Phb2
|
prohibitin 2 |
| chr3_+_154507035 | 0.03 |
ENSRNOT00000017265
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr1_+_199248470 | 0.03 |
ENSRNOT00000025933
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_-_154170409 | 0.03 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chrX_+_157759624 | 0.03 |
ENSRNOT00000003192
|
LOC686087
|
similar to motile sperm domain containing 1 |
| chr4_-_114853868 | 0.03 |
ENSRNOT00000090828
|
Wdr54
|
WD repeat domain 54 |
| chr1_+_97712177 | 0.03 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr3_+_61658245 | 0.03 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr2_-_188745144 | 0.03 |
ENSRNOT00000055533
|
Cks1b
|
CDC28 protein kinase regulatory subunit 1B |
| chr14_+_84465515 | 0.02 |
ENSRNOT00000035591
|
Osm
|
oncostatin M |
| chr13_-_80703615 | 0.02 |
ENSRNOT00000004599
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr11_+_74014983 | 0.02 |
ENSRNOT00000040884
|
NEWGENE_2724
|
glycoprotein V platelet |
| chr15_-_14510828 | 0.02 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr8_-_49075892 | 0.02 |
ENSRNOT00000082687
|
Arcn1
|
archain 1 |
| chr1_-_222178725 | 0.02 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr4_+_162437089 | 0.02 |
ENSRNOT00000038801
|
Clec2d2
|
C-type lectin domain family 2 member D2 |
| chr2_+_211050360 | 0.02 |
ENSRNOT00000026928
|
Psma5
|
proteasome subunit alpha 5 |
| chr10_-_85289777 | 0.02 |
ENSRNOT00000055427
ENSRNOT00000014863 |
Gpr179
|
G protein-coupled receptor 179 |
| chr20_-_3751994 | 0.02 |
ENSRNOT00000072288
|
Pou5f1
|
POU class 5 homeobox 1 |
| chr8_+_55603968 | 0.02 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr13_-_85622314 | 0.02 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr10_+_86399827 | 0.02 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr10_-_90312386 | 0.02 |
ENSRNOT00000028445
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr20_-_3374344 | 0.02 |
ENSRNOT00000082999
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr20_-_5533448 | 0.02 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr4_-_113988246 | 0.02 |
ENSRNOT00000013128
|
LOC103692166
|
WD repeat-containing protein 54 |
| chr8_+_117455262 | 0.02 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr2_+_189582991 | 0.02 |
ENSRNOT00000022627
|
Rab13
|
RAB13, member RAS oncogene family |
| chr3_-_161272460 | 0.02 |
ENSRNOT00000020740
|
Acot8
|
acyl-CoA thioesterase 8 |
| chr20_-_12938891 | 0.02 |
ENSRNOT00000017141
|
RGD1564149
|
similar to Protein C21orf58 |
| chr1_+_47942500 | 0.02 |
ENSRNOT00000025936
|
Wtap
|
Wilms tumor 1 associated protein |
| chr11_-_27971359 | 0.02 |
ENSRNOT00000085629
ENSRNOT00000051060 ENSRNOT00000042581 ENSRNOT00000050073 ENSRNOT00000081066 |
Grik1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr3_+_176217128 | 0.02 |
ENSRNOT00000013609
|
Gid8
|
GID complex subunit 8 |
| chr1_-_89543967 | 0.02 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr15_-_80713153 | 0.02 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr1_+_100470722 | 0.02 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr2_+_44096061 | 0.02 |
ENSRNOT00000018438
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr5_-_24320786 | 0.02 |
ENSRNOT00000061569
|
Ndufaf6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr20_-_5533600 | 0.02 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr14_+_77079402 | 0.02 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr1_-_98486976 | 0.01 |
ENSRNOT00000024083
|
Etfb
|
electron transfer flavoprotein beta subunit |
| chr3_+_80021440 | 0.01 |
ENSRNOT00000018620
|
Acp2
|
acid phosphatase 2, lysosomal |
| chr7_-_127081704 | 0.01 |
ENSRNOT00000055893
|
LOC108351584
|
uncharacterized LOC108351584 |
| chr7_+_12798868 | 0.01 |
ENSRNOT00000038553
|
Prss57
|
protease, serine, 57 |
| chr1_-_8878136 | 0.01 |
ENSRNOT00000064836
ENSRNOT00000075850 |
Vta1
|
vesicle trafficking 1 |
| chr19_-_20508711 | 0.01 |
ENSRNOT00000032408
|
LOC680913
|
hypothetical protein LOC680913 |
| chr5_-_102743417 | 0.01 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr6_-_52401853 | 0.01 |
ENSRNOT00000075513
|
Cdhr3
|
cadherin-related family member 3 |
| chr13_+_99004942 | 0.01 |
ENSRNOT00000085390
|
Acbd3
|
acyl-CoA binding domain containing 3 |
| chr1_-_224986291 | 0.01 |
ENSRNOT00000026284
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr15_+_108286453 | 0.01 |
ENSRNOT00000016986
|
Ubac2
|
UBA domain containing 2 |
| chr12_+_25561777 | 0.01 |
ENSRNOT00000002032
|
Rcc1l
|
RCC1 like |
| chr7_+_118490345 | 0.01 |
ENSRNOT00000046227
|
LOC108348145
|
zinc finger protein 7-like |
| chr13_+_89774764 | 0.01 |
ENSRNOT00000005619
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr4_-_176381477 | 0.01 |
ENSRNOT00000048367
|
Slco1a6
|
solute carrier organic anion transporter family, member 1a6 |
| chr1_-_252116971 | 0.01 |
ENSRNOT00000035013
|
Lipo1
|
lipase, member O1 |
| chr2_+_243656579 | 0.01 |
ENSRNOT00000036993
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr6_+_123034304 | 0.01 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr3_+_141068023 | 0.01 |
ENSRNOT00000036527
|
Kiz
|
kizuna centrosomal protein |
| chr7_-_14054639 | 0.01 |
ENSRNOT00000039852
|
Ilvbl
|
ilvB acetolactate synthase like |
| chr1_+_204861566 | 0.01 |
ENSRNOT00000023214
|
Fam175b
|
family with sequence similarity 175, member B |
| chr19_-_37675077 | 0.01 |
ENSRNOT00000031033
|
Enkd1
|
enkurin domain containing 1 |
| chr2_+_27365148 | 0.01 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr15_-_33218456 | 0.01 |
ENSRNOT00000017753
|
Ajuba
|
ajuba LIM protein |
| chr4_+_118160147 | 0.01 |
ENSRNOT00000022014
|
Fam136a
|
family with sequence similarity 136, member A |
| chr9_+_79659251 | 0.01 |
ENSRNOT00000021656
|
Xrcc5
|
X-ray repair cross complementing 5 |
| chr2_+_243701962 | 0.01 |
ENSRNOT00000016891
|
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr1_-_80617057 | 0.01 |
ENSRNOT00000080453
|
Apoe
|
apolipoprotein E |
| chr7_-_117070936 | 0.01 |
ENSRNOT00000048446
|
Fam83h
|
family with sequence similarity 83, member H |
| chr1_-_206282575 | 0.01 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr9_+_46996156 | 0.01 |
ENSRNOT00000019673
|
Il1r1
|
interleukin 1 receptor type 1 |
| chr9_-_81879211 | 0.01 |
ENSRNOT00000085140
|
Rnf25
|
ring finger protein 25 |
| chr9_+_42620006 | 0.01 |
ENSRNOT00000019966
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr12_-_37699616 | 0.01 |
ENSRNOT00000077803
|
RGD1563482
|
similar to hypothetical protein FLJ38663 |
| chr1_+_215628785 | 0.00 |
ENSRNOT00000054864
|
Lsp1
|
lymphocyte-specific protein 1 |
| chr2_+_218834034 | 0.00 |
ENSRNOT00000018367
|
Dph5
|
diphthamide biosynthesis 5 |
| chr7_+_35069814 | 0.00 |
ENSRNOT00000089228
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr15_+_108960562 | 0.00 |
ENSRNOT00000081906
ENSRNOT00000077482 |
Pcca
|
propionyl-CoA carboxylase alpha subunit |
| chr1_+_101884276 | 0.00 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr16_-_81834945 | 0.00 |
ENSRNOT00000037806
|
F7
|
coagulation factor VII |
| chr2_+_20857202 | 0.00 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr3_+_140024043 | 0.00 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
| chrX_-_128268285 | 0.00 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr1_+_101884019 | 0.00 |
ENSRNOT00000028650
|
Tmem143
|
transmembrane protein 143 |
| chr1_+_225129097 | 0.00 |
ENSRNOT00000026938
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr3_-_160922341 | 0.00 |
ENSRNOT00000029206
|
Tp53tg5
|
TP53 target 5 |
| chr9_+_49647257 | 0.00 |
ENSRNOT00000021899
|
Mrps9
|
mitochondrial ribosomal protein S9 |
| chr10_+_43538160 | 0.00 |
ENSRNOT00000089217
|
Cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr6_+_26537707 | 0.00 |
ENSRNOT00000050102
|
Zfp513
|
zinc finger protein 513 |
| chr19_+_49637016 | 0.00 |
ENSRNOT00000016880
|
Bco1
|
beta-carotene oxygenase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0034443 | plasma lipoprotein particle oxidation(GO:0034441) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0035338 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:0070268 | cuticle development(GO:0042335) cornification(GO:0070268) |
| 0.0 | 0.0 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.0 | 0.0 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.0 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |