Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
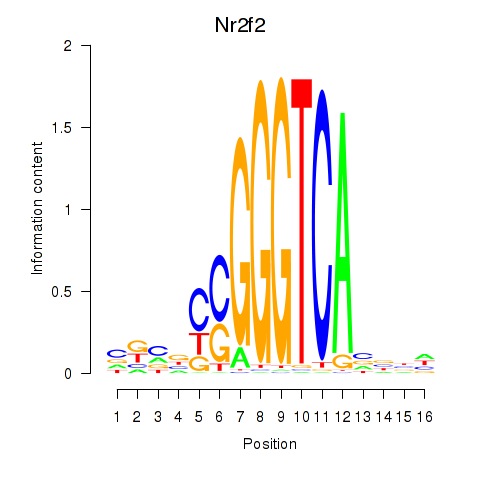
Results for Nr2f2
Z-value: 0.55
Transcription factors associated with Nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f2
|
ENSRNOG00000010308 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f2 | rn6_v1_chr1_-_131454689_131454689 | -0.13 | 8.3e-01 | Click! |
Activity profile of Nr2f2 motif
Sorted Z-values of Nr2f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_943006 | 0.29 |
ENSRNOT00000001449
|
Kl
|
Klotho |
| chrX_+_74315850 | 0.22 |
ENSRNOT00000076538
|
LOC680227
|
LRRGT00193 |
| chr3_-_161131812 | 0.17 |
ENSRNOT00000067008
|
Wfdc11
|
WAP four-disulfide core domain 11 |
| chr1_+_211543751 | 0.17 |
ENSRNOT00000049631
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr3_-_2459385 | 0.15 |
ENSRNOT00000014080
|
Cysrt1
|
cysteine rich tail 1 |
| chr10_+_42933077 | 0.15 |
ENSRNOT00000003361
|
Mfap3
|
microfibrillar-associated protein 3 |
| chr4_+_29639154 | 0.15 |
ENSRNOT00000073687
|
Casd1
|
CAS1 domain containing 1 |
| chr2_-_23289266 | 0.14 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chrX_-_54303729 | 0.14 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr16_-_20807070 | 0.14 |
ENSRNOT00000072536
|
Comp
|
cartilage oligomeric matrix protein |
| chr2_-_178389608 | 0.13 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr18_+_1142782 | 0.13 |
ENSRNOT00000046718
|
Thoc1
|
THO complex 1 |
| chr10_+_88620655 | 0.13 |
ENSRNOT00000055248
|
Hspb9
|
heat shock protein family B (small) member 9 |
| chr17_-_52569036 | 0.12 |
ENSRNOT00000019396
|
Gli3
|
GLI family zinc finger 3 |
| chr1_-_265498831 | 0.11 |
ENSRNOT00000023558
|
Fgf8
|
fibroblast growth factor 8 |
| chr1_+_213595240 | 0.11 |
ENSRNOT00000017137
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr7_-_116106368 | 0.11 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr7_+_141249044 | 0.11 |
ENSRNOT00000084911
|
Aqp5
|
aquaporin 5 |
| chr9_+_81656116 | 0.10 |
ENSRNOT00000083421
|
Slc11a1
|
solute carrier family 11 member 1 |
| chr5_+_105230580 | 0.10 |
ENSRNOT00000010056
|
Acer2
|
alkaline ceramidase 2 |
| chr16_-_32868680 | 0.10 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr1_-_53152866 | 0.10 |
ENSRNOT00000085501
|
Fgfr1op
|
Fgfr1 oncogene partner |
| chr8_+_53678994 | 0.10 |
ENSRNOT00000083419
|
Drd2
|
dopamine receptor D2 |
| chr12_-_41266430 | 0.10 |
ENSRNOT00000001853
ENSRNOT00000081108 |
Oas1b
|
2-5 oligoadenylate synthetase 1B |
| chr11_-_70499200 | 0.10 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chr8_+_53678777 | 0.10 |
ENSRNOT00000045944
|
Drd2
|
dopamine receptor D2 |
| chr15_-_19733967 | 0.10 |
ENSRNOT00000012036
|
Gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr1_+_219481575 | 0.10 |
ENSRNOT00000025507
|
Pold4
|
DNA polymerase delta 4, accessory subunit |
| chr1_+_15620653 | 0.10 |
ENSRNOT00000017401
|
Map7
|
microtubule-associated protein 7 |
| chr11_-_25350974 | 0.09 |
ENSRNOT00000002187
|
Adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr5_-_7941822 | 0.09 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr1_+_31967978 | 0.09 |
ENSRNOT00000081471
ENSRNOT00000021532 |
Trip13
|
thyroid hormone receptor interactor 13 |
| chr7_-_124198200 | 0.09 |
ENSRNOT00000078947
|
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr6_-_88232252 | 0.09 |
ENSRNOT00000047597
|
Rpl10l
|
ribosomal protein L10-like |
| chr10_+_59533480 | 0.09 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr8_-_49106177 | 0.09 |
ENSRNOT00000067558
|
Tmem25
|
transmembrane protein 25 |
| chr1_+_48433079 | 0.08 |
ENSRNOT00000037369
|
Slc22a3
|
solute carrier family 22 member 3 |
| chr5_-_137372524 | 0.08 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr8_+_59769994 | 0.08 |
ENSRNOT00000020309
|
Fbxo22
|
F-box protein 22 |
| chr15_+_34270648 | 0.08 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr17_-_66511870 | 0.08 |
ENSRNOT00000044662
|
Lgals8
|
galectin 8 |
| chr5_-_137372993 | 0.08 |
ENSRNOT00000092823
|
Tmem125
|
transmembrane protein 125 |
| chr3_-_2770620 | 0.08 |
ENSRNOT00000008253
|
Traf2
|
Tnf receptor-associated factor 2 |
| chr2_-_22798214 | 0.08 |
ENSRNOT00000016135
|
Papd4
|
poly(A) RNA polymerase D4, non-canonical |
| chr1_-_239265997 | 0.08 |
ENSRNOT00000037500
|
RGD1359158
|
similar to RIKEN cDNA 1110059E24 |
| chr5_-_169658875 | 0.08 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr2_+_242882306 | 0.08 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr19_+_26173771 | 0.08 |
ENSRNOT00000005581
|
Fbxw9
|
F-box and WD repeat domain containing 9 |
| chr7_+_61236037 | 0.07 |
ENSRNOT00000009776
|
Il22
|
interleukin 22 |
| chrX_-_14910727 | 0.07 |
ENSRNOT00000085079
ENSRNOT00000030146 |
Ssx1
|
SSX family member 1 |
| chr7_+_121930615 | 0.07 |
ENSRNOT00000091270
ENSRNOT00000033975 |
Tnrc6b
|
trinucleotide repeat containing 6B |
| chr2_-_219741886 | 0.07 |
ENSRNOT00000085122
|
Slc35a3
|
solute carrier family 35 member A3 |
| chr7_-_24667301 | 0.07 |
ENSRNOT00000009154
|
Tmem263
|
transmembrane protein 263 |
| chr6_-_133716847 | 0.07 |
ENSRNOT00000072399
|
Rtl1
|
retrotransposon-like 1 |
| chr1_-_192088520 | 0.07 |
ENSRNOT00000047420
|
Palb2
|
partner and localizer of BRCA2 |
| chr7_+_24638208 | 0.07 |
ENSRNOT00000009135
|
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr5_+_4982348 | 0.07 |
ENSRNOT00000010369
|
Lactb2
|
lactamase, beta 2 |
| chr7_-_2786856 | 0.07 |
ENSRNOT00000047530
ENSRNOT00000086939 |
Coq10a
|
coenzyme Q10A |
| chr3_+_151310598 | 0.07 |
ENSRNOT00000092194
|
Mmp24
|
matrix metallopeptidase 24 |
| chr2_+_58534476 | 0.07 |
ENSRNOT00000077646
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr4_+_28441301 | 0.06 |
ENSRNOT00000067087
|
Vps50
|
VPS50 EARP/GARPII complex subunit |
| chr1_-_70235091 | 0.06 |
ENSRNOT00000075745
|
Apeg3
|
antisense paternally expressed gene 3 |
| chr2_-_58534211 | 0.06 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chr4_+_84423653 | 0.06 |
ENSRNOT00000012655
|
Chn2
|
chimerin 2 |
| chr3_-_9326993 | 0.06 |
ENSRNOT00000090137
|
Lamc3
|
laminin subunit gamma 3 |
| chr17_-_21705773 | 0.06 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr12_-_1816414 | 0.06 |
ENSRNOT00000041155
ENSRNOT00000067448 |
Insr
|
insulin receptor |
| chr5_+_169659188 | 0.06 |
ENSRNOT00000066623
|
Nphp4
|
nephrocystin 4 |
| chr13_-_82298404 | 0.06 |
ENSRNOT00000087487
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr18_-_40586260 | 0.06 |
ENSRNOT00000004942
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr20_-_7482747 | 0.06 |
ENSRNOT00000038195
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr11_-_70753632 | 0.06 |
ENSRNOT00000002432
|
Snx4
|
sorting nexin 4 |
| chr12_+_22259713 | 0.06 |
ENSRNOT00000001913
|
Pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr8_+_48422036 | 0.06 |
ENSRNOT00000036051
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr5_-_173425611 | 0.06 |
ENSRNOT00000027060
|
B3galt6
|
Beta-1,3-galactosyltransferase 6 |
| chr10_+_108480368 | 0.06 |
ENSRNOT00000054987
|
Slc26a11
|
solute carrier family 26 member 11 |
| chr10_-_56558487 | 0.06 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr10_-_47018537 | 0.06 |
ENSRNOT00000068351
ENSRNOT00000080083 |
Top3a
|
topoisomerase (DNA) III alpha |
| chr8_+_116343096 | 0.06 |
ENSRNOT00000022092
|
Ifrd2
|
interferon-related developmental regulator 2 |
| chr11_-_51202703 | 0.06 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr14_+_2069409 | 0.06 |
ENSRNOT00000032056
|
Dgkq
|
diacylglycerol kinase, theta |
| chr2_-_116161998 | 0.06 |
ENSRNOT00000012560
|
Gpr160
|
G protein-coupled receptor 160 |
| chr2_-_9504134 | 0.06 |
ENSRNOT00000076996
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr5_+_150957001 | 0.06 |
ENSRNOT00000017549
|
Rpa2
|
replication protein A2 |
| chr19_-_39300893 | 0.05 |
ENSRNOT00000027695
|
Terf2
|
telomeric repeat binding factor 2 |
| chr11_+_83883879 | 0.05 |
ENSRNOT00000050927
|
LOC108348101
|
chloride channel protein 2 |
| chr9_+_17163354 | 0.05 |
ENSRNOT00000026049
|
Polh
|
DNA polymerase eta |
| chr8_-_107602263 | 0.05 |
ENSRNOT00000017658
|
Esyt3
|
extended synaptotagmin 3 |
| chr10_+_46972038 | 0.05 |
ENSRNOT00000023838
|
Mief2
|
mitochondrial elongation factor 2 |
| chr1_+_262905570 | 0.05 |
ENSRNOT00000090765
|
Kcnip2
|
potassium voltage-gated channel interacting protein 2 |
| chr7_-_143863186 | 0.05 |
ENSRNOT00000017096
|
Rarg
|
retinoic acid receptor, gamma |
| chr10_-_15155412 | 0.05 |
ENSRNOT00000026536
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr13_+_51229890 | 0.05 |
ENSRNOT00000060669
|
Cyb5r1
|
cytochrome b5 reductase 1 |
| chr10_+_65566171 | 0.05 |
ENSRNOT00000015651
|
Spag5
|
sperm associated antigen 5 |
| chr9_-_81586116 | 0.05 |
ENSRNOT00000089952
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr2_-_210991509 | 0.05 |
ENSRNOT00000087329
|
Cyb561d1
|
cytochrome b561 family, member D1 |
| chr15_+_3754262 | 0.05 |
ENSRNOT00000035100
|
NEWGENE_1304700
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr11_+_83884048 | 0.05 |
ENSRNOT00000002376
|
LOC108348101
|
chloride channel protein 2 |
| chr9_-_19372673 | 0.05 |
ENSRNOT00000073667
ENSRNOT00000079517 |
Clic5
|
chloride intracellular channel 5 |
| chr15_-_45927804 | 0.05 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr10_-_63219309 | 0.05 |
ENSRNOT00000076357
|
Blmh
|
bleomycin hydrolase |
| chr9_-_92435363 | 0.05 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr11_+_82862695 | 0.05 |
ENSRNOT00000071707
ENSRNOT00000073435 |
Clcn2
|
chloride voltage-gated channel 2 |
| chr10_+_36098051 | 0.05 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chrX_+_43626480 | 0.05 |
ENSRNOT00000068078
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr5_-_134348995 | 0.05 |
ENSRNOT00000015961
|
Faah
|
fatty acid amide hydrolase |
| chr3_-_24601063 | 0.05 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr3_-_7422738 | 0.05 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr5_-_147691997 | 0.05 |
ENSRNOT00000011887
|
Tssk3
|
testis-specific serine kinase 3 |
| chr2_+_17616401 | 0.05 |
ENSRNOT00000064991
|
Edil3
|
EGF like repeats and discoidin domains 3 |
| chr3_+_155297566 | 0.05 |
ENSRNOT00000021435
ENSRNOT00000084866 |
Dhx35
|
DEAH-box helicase 35 |
| chr4_+_2053712 | 0.05 |
ENSRNOT00000045086
|
Rnf32
|
ring finger protein 32 |
| chr20_-_12855891 | 0.05 |
ENSRNOT00000084053
|
Lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr1_+_211582077 | 0.05 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr4_-_79989572 | 0.05 |
ENSRNOT00000013264
ENSRNOT00000086453 |
Dfna5
|
deafness, autosomal dominant 5 (human) |
| chr1_+_102414625 | 0.05 |
ENSRNOT00000089488
|
Kcnc1
|
potassium voltage-gated channel subfamily C member 1 |
| chr4_+_71740532 | 0.05 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
| chr12_-_44174583 | 0.05 |
ENSRNOT00000001490
|
Tesc
|
tescalcin |
| chr7_+_120345180 | 0.05 |
ENSRNOT00000036357
|
Micall1
|
MICAL-like 1 |
| chr14_+_48726045 | 0.04 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr8_-_58542844 | 0.04 |
ENSRNOT00000012041
|
Elmod1
|
ELMO domain containing 1 |
| chr1_-_265560386 | 0.04 |
ENSRNOT00000048592
|
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr5_-_122746839 | 0.04 |
ENSRNOT00000032126
|
Slc35d1
|
solute carrier family 35 member D1 |
| chr12_-_11144754 | 0.04 |
ENSRNOT00000084637
|
Zfp655
|
zinc finger protein 655 |
| chr5_+_63056089 | 0.04 |
ENSRNOT00000081090
|
Tgfbr1
|
transforming growth factor, beta receptor 1 |
| chr18_-_63185510 | 0.04 |
ENSRNOT00000024632
|
Afg3l2
|
AFG3 like matrix AAA peptidase subunit 2 |
| chr9_-_64096265 | 0.04 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr5_-_105582375 | 0.04 |
ENSRNOT00000083373
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr7_-_143217535 | 0.04 |
ENSRNOT00000088316
|
Kb15
|
type II keratin Kb15 |
| chr3_-_159802952 | 0.04 |
ENSRNOT00000011610
|
Oser1
|
oxidative stress responsive serine-rich 1 |
| chr5_+_158090173 | 0.04 |
ENSRNOT00000088766
ENSRNOT00000079516 ENSRNOT00000092026 |
Tas1r2
|
taste 1 receptor member 2 |
| chr1_+_101541266 | 0.04 |
ENSRNOT00000028436
|
Hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr17_-_69862110 | 0.04 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr7_+_127081978 | 0.04 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr7_-_140454268 | 0.04 |
ENSRNOT00000081468
|
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr5_+_154310453 | 0.04 |
ENSRNOT00000013322
|
Gale
|
UDP-galactose-4-epimerase |
| chr1_-_87825333 | 0.04 |
ENSRNOT00000072689
|
Zfp74
|
zinc finger protein 74 |
| chr13_-_73056765 | 0.04 |
ENSRNOT00000000049
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr5_+_169213115 | 0.04 |
ENSRNOT00000013535
|
Nol9
|
nucleolar protein 9 |
| chr4_-_34282351 | 0.04 |
ENSRNOT00000011123
|
Rpa3
|
replication protein A3 |
| chr1_-_89539210 | 0.04 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr2_+_98252925 | 0.04 |
ENSRNOT00000011579
|
Pex2
|
peroxisomal biogenesis factor 2 |
| chr6_+_86713803 | 0.04 |
ENSRNOT00000005861
|
Fam179b
|
family with sequence similarity 179, member B |
| chr11_-_70618347 | 0.04 |
ENSRNOT00000002435
|
Zfp148
|
zinc finger protein 148 |
| chr7_+_143749221 | 0.04 |
ENSRNOT00000014807
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr12_+_9034308 | 0.04 |
ENSRNOT00000001248
|
Flt1
|
FMS-related tyrosine kinase 1 |
| chr19_-_56054142 | 0.04 |
ENSRNOT00000051945
|
Vps9d1
|
VPS9 domain containing 1 |
| chr14_+_77322012 | 0.04 |
ENSRNOT00000088600
ENSRNOT00000041639 |
Zbtb49
|
zinc finger and BTB domain containing 49 |
| chr3_+_150052483 | 0.04 |
ENSRNOT00000022265
|
RGD1561517
|
similar to chromosome 20 open reading frame 144 |
| chr4_-_34779776 | 0.04 |
ENSRNOT00000077329
|
Ica1
|
islet cell autoantigen 1 |
| chr10_+_15485905 | 0.04 |
ENSRNOT00000027609
|
Tmem8a
|
transmembrane protein 8A |
| chr2_-_21931720 | 0.04 |
ENSRNOT00000018449
|
Msh3
|
mutS homolog 3 |
| chr1_+_211544157 | 0.04 |
ENSRNOT00000023610
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr4_-_34780193 | 0.04 |
ENSRNOT00000011651
|
Ica1
|
islet cell autoantigen 1 |
| chr10_-_78219793 | 0.04 |
ENSRNOT00000003369
|
Tom1l1
|
target of myb1 like 1 membrane trafficking protein |
| chr9_+_16647922 | 0.04 |
ENSRNOT00000031625
|
Klc4
|
kinesin light chain 4 |
| chr20_+_3823596 | 0.04 |
ENSRNOT00000087670
|
Rxrb
|
retinoid X receptor beta |
| chr2_-_195678848 | 0.04 |
ENSRNOT00000028303
ENSRNOT00000075569 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr1_+_219833299 | 0.04 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr15_+_3938075 | 0.04 |
ENSRNOT00000065644
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_+_95397991 | 0.04 |
ENSRNOT00000039649
|
Zfp939
|
zinc finger protein 939 |
| chr11_-_88016825 | 0.04 |
ENSRNOT00000079595
|
Hic2
|
HIC ZBTB transcriptional repressor 2 |
| chr7_+_142106465 | 0.04 |
ENSRNOT00000042786
ENSRNOT00000088681 |
Letmd1
|
LETM1 domain containing 1 |
| chr7_-_145575981 | 0.04 |
ENSRNOT00000074622
|
LOC102555445
|
16.5 kDa submandibular gland glycoprotein-like |
| chr8_-_23148396 | 0.03 |
ENSRNOT00000075237
|
Acp5
|
acid phosphatase 5, tartrate resistant |
| chr5_+_140692779 | 0.03 |
ENSRNOT00000019101
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr5_-_58183017 | 0.03 |
ENSRNOT00000020982
|
Ccl19
|
C-C motif chemokine ligand 19 |
| chr16_+_81587446 | 0.03 |
ENSRNOT00000092680
|
NEWGENE_1582994
|
DCN1, defective in cullin neddylation 1, domain containing 2 |
| chr7_-_120967490 | 0.03 |
ENSRNOT00000046399
|
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr15_-_46719252 | 0.03 |
ENSRNOT00000015452
|
Mtmr9
|
myotubularin related protein 9 |
| chr1_+_29413365 | 0.03 |
ENSRNOT00000019047
ENSRNOT00000088885 |
Hint3
|
histidine triad nucleotide binding protein 3 |
| chr13_+_82298534 | 0.03 |
ENSRNOT00000003636
|
Mettl18
|
methyltransferase like 18 |
| chr5_-_160156209 | 0.03 |
ENSRNOT00000045237
|
Fblim1
|
filamin binding LIM protein 1 |
| chr15_+_34296922 | 0.03 |
ENSRNOT00000026430
|
Rec8
|
REC8 meiotic recombination protein |
| chr1_-_265798167 | 0.03 |
ENSRNOT00000079483
|
Ldb1
|
LIM domain binding 1 |
| chr17_+_18031228 | 0.03 |
ENSRNOT00000081708
|
Tpmt
|
thiopurine S-methyltransferase |
| chr20_+_4369394 | 0.03 |
ENSRNOT00000088734
ENSRNOT00000091491 |
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr8_+_48569328 | 0.03 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr3_+_145032200 | 0.03 |
ENSRNOT00000068210
|
Syndig1
|
synapse differentiation inducing 1 |
| chr13_-_89654244 | 0.03 |
ENSRNOT00000004896
|
Ppox
|
protoporphyrinogen oxidase |
| chr1_-_205842428 | 0.03 |
ENSRNOT00000024655
|
Dhx32
|
DEAH-box helicase 32 (putative) |
| chr3_-_118137125 | 0.03 |
ENSRNOT00000012287
ENSRNOT00000084154 |
Cops2
|
COP9 signalosome subunit 2 |
| chr3_+_161425988 | 0.03 |
ENSRNOT00000065184
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr10_-_62191512 | 0.03 |
ENSRNOT00000090262
|
Rpa1
|
replication protein A1 |
| chr14_+_39154529 | 0.03 |
ENSRNOT00000003191
|
Gabra4
|
gamma-aminobutyric acid type A receptor alpha4 subunit |
| chr6_-_99400822 | 0.03 |
ENSRNOT00000089594
|
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr5_-_140015541 | 0.03 |
ENSRNOT00000016410
|
Zmpste24
|
zinc metallopeptidase STE24 |
| chr10_+_93811505 | 0.03 |
ENSRNOT00000081937
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr2_+_112868707 | 0.03 |
ENSRNOT00000017805
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr1_-_277902279 | 0.03 |
ENSRNOT00000078416
|
Ablim1
|
actin-binding LIM protein 1 |
| chr19_+_38846201 | 0.03 |
ENSRNOT00000030768
ENSRNOT00000063977 |
Tango6
|
transport and golgi organization 6 homolog |
| chr11_+_73079168 | 0.03 |
ENSRNOT00000080834
|
Acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr6_+_44009872 | 0.03 |
ENSRNOT00000082657
|
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr2_-_252382955 | 0.03 |
ENSRNOT00000021705
|
Rpf1
|
ribosome production factor 1 homolog |
| chr14_-_44348691 | 0.03 |
ENSRNOT00000030485
|
Ube2k
|
ubiquitin-conjugating enzyme E2K |
| chr11_-_72378895 | 0.03 |
ENSRNOT00000058885
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr19_+_11473541 | 0.03 |
ENSRNOT00000082066
|
Amfr
|
autocrine motility factor receptor |
| chr16_-_55002322 | 0.03 |
ENSRNOT00000036092
|
Zdhhc2
|
zinc finger, DHHC-type containing 2 |
| chr15_-_45376476 | 0.03 |
ENSRNOT00000065838
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr1_+_199449973 | 0.03 |
ENSRNOT00000029994
|
Trim72
|
tripartite motif containing 72 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051944 | glial cell-derived neurotrophic factor secretion(GO:0044467) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) negative regulation of dopamine receptor signaling pathway(GO:0060160) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.1 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.0 | 0.1 | GO:0048382 | mesendoderm development(GO:0048382) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.3 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.1 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.0 | 0.1 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.0 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.0 | GO:0003343 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.0 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.0 | GO:0032919 | spermine acetylation(GO:0032919) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:0090292 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.0 | GO:0001582 | detection of chemical stimulus involved in sensory perception of sweet taste(GO:0001582) |
| 0.0 | 0.0 | GO:0071283 | cellular response to iron(III) ion(GO:0071283) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:1903767 | sweet taste receptor complex(GO:1903767) taste receptor complex(GO:1903768) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.0 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.0 | GO:0000250 | lanosterol synthase activity(GO:0000250) oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity(GO:0003842) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.0 | GO:0032142 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.0 | 0.0 | GO:0015203 | ornithine decarboxylase inhibitor activity(GO:0008073) polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.0 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |