Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
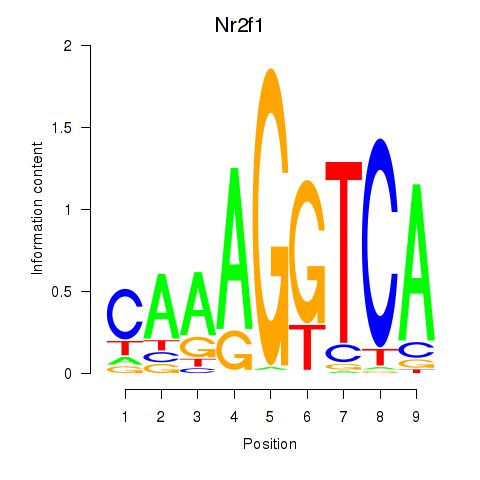
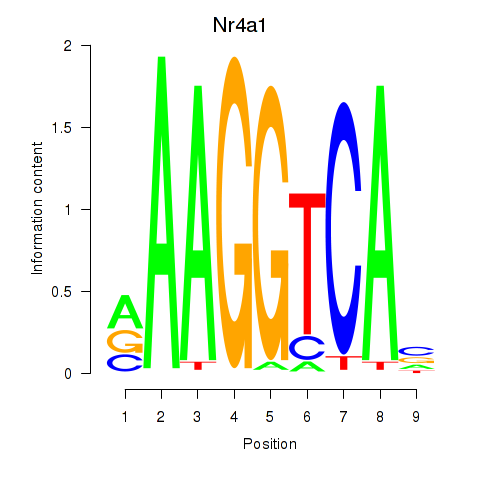
Results for Nr2f1_Nr4a1
Z-value: 0.74
Transcription factors associated with Nr2f1_Nr4a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f1
|
ENSRNOG00000014795 | nuclear receptor subfamily 2, group F, member 1 |
|
Nr4a1
|
ENSRNOG00000007607 | nuclear receptor subfamily 4, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f1 | rn6_v1_chr2_-_5577369_5577369 | -0.50 | 3.9e-01 | Click! |
| Nr4a1 | rn6_v1_chr7_+_142905758_142905758 | -0.35 | 5.6e-01 | Click! |
Activity profile of Nr2f1_Nr4a1 motif
Sorted Z-values of Nr2f1_Nr4a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_123106694 | 0.70 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr14_+_43694183 | 0.65 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chr18_-_26211445 | 0.40 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr3_+_147609095 | 0.38 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr17_+_15845931 | 0.32 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
| chr1_+_101412736 | 0.31 |
ENSRNOT00000067468
|
Lhb
|
luteinizing hormone beta polypeptide |
| chr1_+_242572533 | 0.29 |
ENSRNOT00000035123
|
Tmem252
|
transmembrane protein 252 |
| chr12_+_47024442 | 0.28 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr8_+_118378059 | 0.26 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr10_-_40375605 | 0.26 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr1_+_31264755 | 0.26 |
ENSRNOT00000028988
|
LOC679739
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr4_+_78263866 | 0.25 |
ENSRNOT00000033807
|
AI854703
|
expressed sequence AI854703 |
| chr8_-_23014499 | 0.25 |
ENSRNOT00000017820
|
Ccdc151
|
coiled-coil domain containing 151 |
| chr4_+_170149029 | 0.23 |
ENSRNOT00000073287
|
LOC103690002
|
histone H2A.J |
| chr5_+_50381244 | 0.23 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr5_-_166924136 | 0.22 |
ENSRNOT00000085251
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr10_-_71849293 | 0.22 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chr12_-_40227297 | 0.21 |
ENSRNOT00000030600
|
Fam109a
|
family with sequence similarity 109, member A |
| chr7_-_121943121 | 0.21 |
ENSRNOT00000046460
|
Rpl26-ps1
|
ribosomal protein L26, pseudogene 1 |
| chr1_+_267607416 | 0.21 |
ENSRNOT00000087894
ENSRNOT00000076367 ENSRNOT00000016851 |
Gsto2
Gsto1
|
glutathione S-transferase omega 2 glutathione S-transferase omega 1 |
| chr2_-_140464607 | 0.20 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr1_+_71416110 | 0.20 |
ENSRNOT00000084899
|
Zfp787
|
zinc finger protein 787 |
| chr2_-_119140110 | 0.20 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
| chr1_+_276659542 | 0.20 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr7_+_121311024 | 0.20 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr2_+_204512302 | 0.19 |
ENSRNOT00000021846
|
Casq2
|
calsequestrin 2 |
| chr10_+_110139783 | 0.19 |
ENSRNOT00000054939
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr2_-_123396147 | 0.19 |
ENSRNOT00000079004
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr10_-_97756521 | 0.19 |
ENSRNOT00000045902
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr2_+_187347602 | 0.18 |
ENSRNOT00000025384
|
Nes
|
nestin |
| chr4_-_77706994 | 0.18 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr19_-_37427989 | 0.18 |
ENSRNOT00000022863
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr20_-_5618254 | 0.18 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chrX_-_113584459 | 0.17 |
ENSRNOT00000025923
|
Kcne5
|
potassium voltage-gated channel subfamily E regulatory subunit 5 |
| chr8_+_116094851 | 0.17 |
ENSRNOT00000084120
|
RGD1307461
|
similar to RIKEN cDNA 6430571L13 gene; similar to g20 protein |
| chr6_-_135412312 | 0.17 |
ENSRNOT00000010610
|
Ankrd9
|
ankyrin repeat domain 9 |
| chr8_+_98745310 | 0.17 |
ENSRNOT00000019938
|
Zic4
|
Zic family member 4 |
| chr19_-_25166528 | 0.16 |
ENSRNOT00000007775
|
Rln3
|
relaxin 3 |
| chr4_-_148845267 | 0.16 |
ENSRNOT00000037397
|
Tmem72
|
transmembrane protein 72 |
| chr12_-_30566032 | 0.15 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr7_+_116090189 | 0.15 |
ENSRNOT00000081571
|
Psca
|
prostate stem cell antigen |
| chr20_+_29951637 | 0.15 |
ENSRNOT00000074387
|
LOC100361018
|
rCG22048-like |
| chr1_-_222250980 | 0.15 |
ENSRNOT00000028734
|
Nudt22
|
nudix hydrolase 22 |
| chr2_-_14701903 | 0.15 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr1_-_192057612 | 0.15 |
ENSRNOT00000024471
|
Ndufab1
|
NADH:ubiquinone oxidoreductase subunit AB1 |
| chr20_-_3818045 | 0.15 |
ENSRNOT00000091622
|
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr19_+_52521809 | 0.14 |
ENSRNOT00000081019
|
Klhl36
|
kelch-like family member 36 |
| chr2_+_119139717 | 0.14 |
ENSRNOT00000016051
|
Ndufb5
|
NADH:ubiquinone oxidoreductase subunit B5 |
| chr11_-_59110562 | 0.14 |
ENSRNOT00000047907
ENSRNOT00000042024 |
Lsamp
|
limbic system-associated membrane protein |
| chr10_-_15600858 | 0.14 |
ENSRNOT00000035459
|
Hbq1b
|
hemoglobin, theta 1B |
| chr12_-_44174583 | 0.14 |
ENSRNOT00000001490
|
Tesc
|
tescalcin |
| chr3_-_8766433 | 0.14 |
ENSRNOT00000021865
|
Kyat1
|
kynurenine aminotransferase 1 |
| chr1_-_89488223 | 0.14 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr5_-_134927235 | 0.14 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr10_-_84976170 | 0.14 |
ENSRNOT00000013542
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr8_+_48569328 | 0.13 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr12_-_18526250 | 0.13 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr10_-_15235740 | 0.13 |
ENSRNOT00000027170
|
Mcrip2
|
MAPK regulated co-repressor interacting protein 2 |
| chr8_+_59900651 | 0.13 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chr7_+_35125424 | 0.13 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr7_-_12432130 | 0.13 |
ENSRNOT00000077301
|
Atp5d
|
ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit |
| chr19_+_10596960 | 0.13 |
ENSRNOT00000021769
|
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr8_-_23000455 | 0.13 |
ENSRNOT00000091027
ENSRNOT00000017493 |
Rgl3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr5_-_168004724 | 0.13 |
ENSRNOT00000024711
|
Park7
|
Parkinsonism associated deglycase |
| chr6_-_136145837 | 0.13 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr5_-_154438361 | 0.13 |
ENSRNOT00000085003
|
AC141344.2
|
|
| chr14_+_84237520 | 0.12 |
ENSRNOT00000083580
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr9_+_61692154 | 0.12 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr10_-_38782419 | 0.12 |
ENSRNOT00000073964
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr14_-_84937725 | 0.12 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr7_-_143793774 | 0.12 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr3_+_2480232 | 0.12 |
ENSRNOT00000014489
|
Tprn
|
taperin |
| chr18_-_16542165 | 0.12 |
ENSRNOT00000079381
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr11_+_30904733 | 0.12 |
ENSRNOT00000036027
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr7_-_127081704 | 0.12 |
ENSRNOT00000055893
|
LOC108351584
|
uncharacterized LOC108351584 |
| chr1_-_31528083 | 0.12 |
ENSRNOT00000061041
|
Rpl26-ps2
|
ribosomal protein L26, pseudogene 2 |
| chr1_-_24193567 | 0.12 |
ENSRNOT00000040736
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr19_-_54245855 | 0.12 |
ENSRNOT00000023855
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr4_-_82133605 | 0.12 |
ENSRNOT00000008023
|
NEWGENE_2813
|
homeobox A2 |
| chr2_-_225107283 | 0.12 |
ENSRNOT00000055711
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr15_+_2631529 | 0.12 |
ENSRNOT00000018897
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr5_+_151436464 | 0.11 |
ENSRNOT00000012463
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr5_+_150032999 | 0.11 |
ENSRNOT00000013301
|
Srsf4
|
serine and arginine rich splicing factor 4 |
| chr2_-_243407608 | 0.11 |
ENSRNOT00000014631
|
Mttp
|
microsomal triglyceride transfer protein |
| chr6_-_44363915 | 0.11 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr4_-_123118186 | 0.11 |
ENSRNOT00000038096
|
LOC100361898
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr9_+_43259709 | 0.11 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr9_-_43127887 | 0.11 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr2_-_149088787 | 0.11 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr2_-_197991574 | 0.11 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr1_-_7480825 | 0.11 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr8_-_130491998 | 0.10 |
ENSRNOT00000064114
|
Higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr3_+_113318563 | 0.10 |
ENSRNOT00000089230
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr9_+_17340341 | 0.10 |
ENSRNOT00000026637
ENSRNOT00000026559 ENSRNOT00000042790 ENSRNOT00000044163 ENSRNOT00000083811 |
Vegfa
|
vascular endothelial growth factor A |
| chr7_+_12229379 | 0.10 |
ENSRNOT00000060811
|
Adamtsl5
|
ADAMTS-like 5 |
| chr6_+_126434226 | 0.10 |
ENSRNOT00000090857
|
Chga
|
chromogranin A |
| chr3_-_148428288 | 0.10 |
ENSRNOT00000072663
|
Dusp15
|
dual specificity phosphatase 15 |
| chr4_+_169161585 | 0.10 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr11_+_72044096 | 0.10 |
ENSRNOT00000034757
|
Senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr10_-_89958805 | 0.10 |
ENSRNOT00000084266
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr3_-_172566010 | 0.10 |
ENSRNOT00000071913
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr4_+_64088900 | 0.10 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr16_+_19097391 | 0.10 |
ENSRNOT00000018018
|
Calr3
|
calreticulin 3 |
| chr6_+_8669722 | 0.09 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr5_-_166926636 | 0.09 |
ENSRNOT00000023287
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr1_-_242083484 | 0.09 |
ENSRNOT00000065921
|
Tjp2
|
tight junction protein 2 |
| chr6_+_23337571 | 0.09 |
ENSRNOT00000011832
|
RGD1304963
|
similar to hypothetical protein MGC38716 |
| chr1_+_89162639 | 0.09 |
ENSRNOT00000028508
|
Atp4a
|
ATPase H+/K+ transporting alpha subunit |
| chr12_-_30268990 | 0.09 |
ENSRNOT00000079546
|
LOC103693015
|
vitamin K epoxide reductase complex subunit 1-like protein 1 |
| chr1_-_84812486 | 0.09 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr6_-_6842758 | 0.09 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr17_+_32904119 | 0.09 |
ENSRNOT00000059854
|
Serpinb1a
|
serpin family B member 1A |
| chr7_-_12311149 | 0.09 |
ENSRNOT00000046886
|
Dazap1
|
DAZ associated protein 1 |
| chr10_-_56429748 | 0.09 |
ENSRNOT00000020675
ENSRNOT00000092704 |
Spem1
|
spermatid maturation 1 |
| chr5_-_75319189 | 0.09 |
ENSRNOT00000047200
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr10_+_74874130 | 0.09 |
ENSRNOT00000076050
ENSRNOT00000044226 ENSRNOT00000085052 ENSRNOT00000076196 |
Sept4
|
septin 4 |
| chr7_-_34485034 | 0.08 |
ENSRNOT00000007351
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr5_-_59149625 | 0.08 |
ENSRNOT00000040641
ENSRNOT00000084216 |
Spag8
|
sperm associated antigen 8 |
| chr19_+_9024451 | 0.08 |
ENSRNOT00000073744
|
LOC100910721
|
60S ribosomal protein L26-like |
| chr7_+_117951154 | 0.08 |
ENSRNOT00000060181
|
Znf7
|
zinc finger protein 7 |
| chr3_+_170252901 | 0.08 |
ENSRNOT00000005871
|
Mc3r
|
melanocortin 3 receptor |
| chrX_-_68562301 | 0.08 |
ENSRNOT00000076720
|
Ophn1
|
oligophrenin 1 |
| chr5_+_173640780 | 0.08 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr20_-_30406295 | 0.08 |
ENSRNOT00000038537
|
Unc5b
|
unc-5 netrin receptor B |
| chr1_+_207508414 | 0.08 |
ENSRNOT00000054896
|
Nps
|
neuropeptide S |
| chr1_-_91042230 | 0.08 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr20_+_4824226 | 0.08 |
ENSRNOT00000001113
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr20_+_5085712 | 0.08 |
ENSRNOT00000083284
|
Abhd16a
|
abhydrolase domain containing 16A |
| chr1_+_224927764 | 0.08 |
ENSRNOT00000050814
|
AC099294.1
|
|
| chr1_-_80630038 | 0.08 |
ENSRNOT00000025281
|
Tomm40
|
translocase of outer mitochondrial membrane 40 |
| chr6_-_106971250 | 0.08 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr10_+_109658032 | 0.08 |
ENSRNOT00000054965
|
Mrpl12
|
mitochondrial ribosomal protein L12 |
| chr2_-_123396386 | 0.08 |
ENSRNOT00000046700
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr9_-_9985630 | 0.08 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr18_+_74156553 | 0.08 |
ENSRNOT00000022892
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr11_-_32088002 | 0.08 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr3_+_61756148 | 0.08 |
ENSRNOT00000002134
|
Mtx2
|
metaxin 2 |
| chr5_+_159484370 | 0.08 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr8_-_118378460 | 0.08 |
ENSRNOT00000047247
|
RGD1563705
|
similar to ribosomal protein S23 |
| chr20_-_4489281 | 0.07 |
ENSRNOT00000031548
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr10_+_56662561 | 0.07 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr9_+_10535340 | 0.07 |
ENSRNOT00000075408
|
Znrf4
|
zinc and ring finger 4 |
| chr20_+_26534535 | 0.07 |
ENSRNOT00000000423
|
Tmem167b
|
transmembrane protein 167B |
| chr16_-_81243757 | 0.07 |
ENSRNOT00000024677
|
Gas6
|
growth arrest specific 6 |
| chr9_-_92291220 | 0.07 |
ENSRNOT00000093357
|
Dner
|
delta/notch-like EGF repeat containing |
| chr5_-_157573183 | 0.07 |
ENSRNOT00000064418
|
Minos1
|
mitochondrial inner membrane organizing system 1 |
| chr9_-_10441834 | 0.07 |
ENSRNOT00000043704
|
Rpl36
|
ribosomal protein L36 |
| chr10_+_56662242 | 0.07 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr7_+_11724962 | 0.07 |
ENSRNOT00000026551
|
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr19_+_54245950 | 0.07 |
ENSRNOT00000024033
|
Cox4i1
|
cytochrome c oxidase subunit 4i1 |
| chr10_-_89959176 | 0.07 |
ENSRNOT00000028264
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr17_-_389967 | 0.07 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr1_+_141550633 | 0.07 |
ENSRNOT00000020047
|
Mesp2
|
mesoderm posterior bHLH transcription factor 2 |
| chr5_-_56576676 | 0.07 |
ENSRNOT00000029712
|
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr11_+_64472072 | 0.07 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr10_-_91448477 | 0.07 |
ENSRNOT00000038836
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr6_+_126170911 | 0.07 |
ENSRNOT00000077477
|
Rin3
|
Ras and Rab interactor 3 |
| chr1_+_213595240 | 0.06 |
ENSRNOT00000017137
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr14_-_79464770 | 0.06 |
ENSRNOT00000008932
|
Grpel1
|
GrpE-like 1, mitochondrial |
| chr7_-_117978787 | 0.06 |
ENSRNOT00000074262
|
Zfp647
|
zinc finger protein 647 |
| chr4_-_100232559 | 0.06 |
ENSRNOT00000016984
|
Vamp5
|
vesicle-associated membrane protein 5 |
| chr15_+_48789165 | 0.06 |
ENSRNOT00000044562
|
Zfp395
|
zinc finger protein 395 |
| chr1_-_262013619 | 0.06 |
ENSRNOT00000021278
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr11_+_37798370 | 0.06 |
ENSRNOT00000002679
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr12_-_23624212 | 0.06 |
ENSRNOT00000064405
ENSRNOT00000001943 |
Rasa4
|
RAS p21 protein activator 4 |
| chr14_-_8548310 | 0.06 |
ENSRNOT00000092436
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr3_+_81287242 | 0.06 |
ENSRNOT00000086530
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr5_-_160179978 | 0.06 |
ENSRNOT00000022820
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr3_-_123702732 | 0.06 |
ENSRNOT00000028859
|
RGD1311739
|
similar to RIKEN cDNA 1700037H04 |
| chr7_-_144960527 | 0.06 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr4_-_130659697 | 0.06 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr9_-_9702306 | 0.06 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr13_-_89518939 | 0.06 |
ENSRNOT00000004228
|
Sdhc
|
succinate dehydrogenase complex subunit C |
| chr6_+_107581608 | 0.06 |
ENSRNOT00000058101
|
Acot6
|
acyl-CoA thioesterase 6 |
| chr4_+_122781095 | 0.06 |
ENSRNOT00000008962
|
Hdac11
|
histone deacetylase 11 |
| chr10_-_45923829 | 0.06 |
ENSRNOT00000035285
|
Olr1462
|
olfactory receptor 1462 |
| chr8_+_117679278 | 0.06 |
ENSRNOT00000042114
|
Uqcrc1
|
ubiquinol-cytochrome c reductase core protein I |
| chr19_-_56677084 | 0.06 |
ENSRNOT00000024084
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr16_-_8350067 | 0.06 |
ENSRNOT00000026932
|
LOC100362432
|
mitochondrial import inner membrane translocase subunit Tim23-like |
| chr1_+_261158261 | 0.06 |
ENSRNOT00000071965
|
Pgam1
|
phosphoglycerate mutase 1 |
| chr2_-_187401786 | 0.06 |
ENSRNOT00000025624
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr8_+_117280705 | 0.06 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr10_+_37724915 | 0.06 |
ENSRNOT00000008477
|
Vdac1
|
voltage-dependent anion channel 1 |
| chr5_+_134927609 | 0.06 |
ENSRNOT00000018506
|
Lrrc41
|
leucine rich repeat containing 41 |
| chr7_+_144052061 | 0.06 |
ENSRNOT00000020103
ENSRNOT00000091378 |
Amhr2
|
anti-Mullerian hormone receptor type 2 |
| chr19_-_14945302 | 0.06 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr12_-_29952196 | 0.06 |
ENSRNOT00000086175
|
Tmem248
|
transmembrane protein 248 |
| chrX_+_110818716 | 0.06 |
ENSRNOT00000086308
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_197813963 | 0.06 |
ENSRNOT00000045531
|
LOC100361060
|
ribosomal protein L36-like |
| chr14_+_60764409 | 0.06 |
ENSRNOT00000005168
|
Lgi2
|
leucine-rich repeat LGI family, member 2 |
| chr7_-_118518193 | 0.06 |
ENSRNOT00000075636
|
LOC102555083
|
zinc finger protein 250-like |
| chrX_-_123662350 | 0.06 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr9_-_81565416 | 0.06 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr7_+_120380544 | 0.06 |
ENSRNOT00000015009
|
Polr2f
|
RNA polymerase II subunit F |
| chr5_-_174921 | 0.06 |
ENSRNOT00000056239
|
Tipinl1
|
TIMELESS interacting protein like 1 |
| chr9_+_20213776 | 0.06 |
ENSRNOT00000071439
|
LOC100911515
|
triosephosphate isomerase-like |
| chr4_+_161696903 | 0.06 |
ENSRNOT00000055889
|
LOC100362909
|
rCG30099-like |
| chr1_-_263762785 | 0.05 |
ENSRNOT00000018221
|
Cpn1
|
carboxypeptidase N subunit 1 |
| chr20_-_3819200 | 0.05 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr10_+_77537340 | 0.05 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f1_Nr4a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.1 | 0.2 | GO:0060066 | oviduct development(GO:0060066) |
| 0.1 | 0.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.4 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.2 | GO:0045105 | intermediate filament polymerization or depolymerization(GO:0045105) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.0 | 0.1 | GO:0045560 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0002339 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.0 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.1 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.0 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0021627 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.1 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.0 | 0.0 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.4 | GO:0030823 | regulation of cGMP metabolic process(GO:0030823) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.0 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.0 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:0045273 | respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 1.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) sperm annulus(GO:0097227) |
| 0.0 | 0.0 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.0 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.2 | GO:0030614 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.0 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |