Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
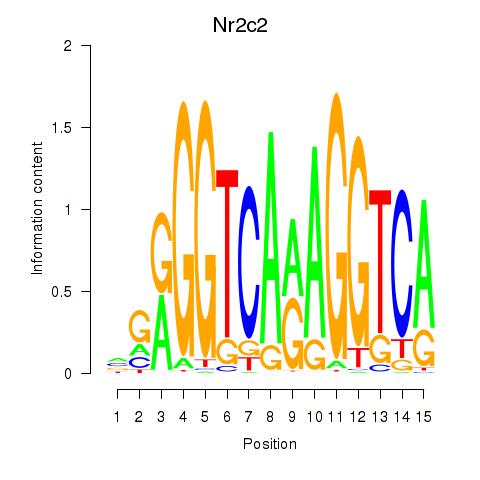
Results for Nr2c2
Z-value: 0.40
Transcription factors associated with Nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2c2
|
ENSRNOG00000010536 | nuclear receptor subfamily 2, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2c2 | rn6_v1_chr4_+_124005280_124005280 | 0.41 | 5.0e-01 | Click! |
Activity profile of Nr2c2 motif
Sorted Z-values of Nr2c2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_13778178 | 0.36 |
ENSRNOT00000058314
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr12_-_18526250 | 0.29 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr10_+_59533480 | 0.28 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr6_+_43001948 | 0.23 |
ENSRNOT00000007374
|
Hpcal1
|
hippocalcin-like 1 |
| chr16_-_20890949 | 0.22 |
ENSRNOT00000081977
|
Homer3
|
homer scaffolding protein 3 |
| chr1_-_226049929 | 0.20 |
ENSRNOT00000007320
|
Best1
|
bestrophin 1 |
| chr16_-_59366824 | 0.19 |
ENSRNOT00000015062
|
RGD1304810
|
similar to 6430573F11Rik protein |
| chr3_-_7498555 | 0.19 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr5_-_156734541 | 0.19 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr2_-_225107283 | 0.16 |
ENSRNOT00000055711
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr4_-_71763679 | 0.16 |
ENSRNOT00000024037
|
Epha1
|
Eph receptor A1 |
| chr5_-_137372993 | 0.16 |
ENSRNOT00000092823
|
Tmem125
|
transmembrane protein 125 |
| chr8_-_80631873 | 0.16 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr1_+_198690794 | 0.16 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr1_+_213886775 | 0.15 |
ENSRNOT00000020864
|
Pkp3
|
plakophilin 3 |
| chr1_-_262013619 | 0.14 |
ENSRNOT00000021278
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chr16_+_7303578 | 0.14 |
ENSRNOT00000025656
|
Sema3g
|
semaphorin 3G |
| chr3_-_153454160 | 0.13 |
ENSRNOT00000010298
|
Ghrh
|
growth hormone releasing hormone |
| chr10_+_86399827 | 0.13 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr5_+_120250616 | 0.13 |
ENSRNOT00000070967
|
Ak4
|
adenylate kinase 4 |
| chr7_+_28715300 | 0.11 |
ENSRNOT00000006760
ENSRNOT00000089161 |
Nup37
|
nucleoporin 37 |
| chr20_+_13662867 | 0.11 |
ENSRNOT00000032257
|
RGD1564162
|
similar to Homo sapiens fetal lung specific expression unknown |
| chr3_+_61658245 | 0.11 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr5_-_137372524 | 0.11 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr10_-_59888198 | 0.10 |
ENSRNOT00000093482
ENSRNOT00000049311 ENSRNOT00000093230 |
Aspa
|
aspartoacylase |
| chr10_+_104437648 | 0.10 |
ENSRNOT00000035001
|
AC130970.1
|
|
| chr8_+_70112925 | 0.10 |
ENSRNOT00000082401
|
Megf11
|
multiple EGF-like-domains 11 |
| chr10_-_94460732 | 0.10 |
ENSRNOT00000014508
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr11_+_32440237 | 0.09 |
ENSRNOT00000040844
|
Kcne2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chr5_+_78483893 | 0.09 |
ENSRNOT00000059183
ENSRNOT00000059181 |
Rgs3
|
regulator of G-protein signaling 3 |
| chr13_-_90602365 | 0.09 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr1_-_43884267 | 0.09 |
ENSRNOT00000024418
|
Cnksr3
|
Cnksr family member 3 |
| chrX_-_123662350 | 0.09 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr7_-_117369159 | 0.09 |
ENSRNOT00000016424
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr7_-_130405347 | 0.09 |
ENSRNOT00000013985
|
Cpt1b
|
carnitine palmitoyltransferase 1B |
| chr12_-_30566032 | 0.08 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr19_-_46101250 | 0.08 |
ENSRNOT00000015874
|
Adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr1_+_72784966 | 0.08 |
ENSRNOT00000090065
ENSRNOT00000041527 |
Tmem86b
|
transmembrane protein 86B |
| chr10_-_97754445 | 0.08 |
ENSRNOT00000088215
|
Slc16a6
|
solute carrier family 16, member 6 |
| chrX_+_45420596 | 0.08 |
ENSRNOT00000051897
|
Sts
|
steroid sulfatase (microsomal), isozyme S |
| chr1_-_89543967 | 0.08 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr3_-_2938883 | 0.08 |
ENSRNOT00000084196
|
LOC108348105
|
allergen Fel d 4-like |
| chr13_-_102721218 | 0.07 |
ENSRNOT00000005459
|
Marc1
|
mitochondrial amidoxime reducing component 1 |
| chr20_-_5123073 | 0.07 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr7_+_12471824 | 0.07 |
ENSRNOT00000068197
|
Sbno2
|
strawberry notch homolog 2 |
| chr10_-_103873246 | 0.07 |
ENSRNOT00000039387
|
Ush1g
|
Usher syndrome 1G |
| chr7_-_127081704 | 0.07 |
ENSRNOT00000055893
|
LOC108351584
|
uncharacterized LOC108351584 |
| chr6_-_132608600 | 0.07 |
ENSRNOT00000015855
|
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr3_+_149424392 | 0.07 |
ENSRNOT00000044574
|
Bpifb4
|
BPI fold containing family B, member 4 |
| chrX_+_15598652 | 0.07 |
ENSRNOT00000082383
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr20_-_14193690 | 0.07 |
ENSRNOT00000058237
|
Upb1
|
beta-ureidopropionase 1 |
| chr5_+_137243094 | 0.07 |
ENSRNOT00000067237
|
Med8
|
mediator complex subunit 8 |
| chr5_+_70441123 | 0.07 |
ENSRNOT00000087517
|
Fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr8_+_130762674 | 0.07 |
ENSRNOT00000005422
|
Snrk
|
SNF related kinase |
| chr3_+_101051955 | 0.06 |
ENSRNOT00000007681
|
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4 |
| chr3_-_1584946 | 0.06 |
ENSRNOT00000031058
|
Pax8
|
paired box 8 |
| chr6_+_10348308 | 0.06 |
ENSRNOT00000034991
|
Epas1
|
endothelial PAS domain protein 1 |
| chr1_-_226791773 | 0.06 |
ENSRNOT00000082482
ENSRNOT00000065376 ENSRNOT00000054812 ENSRNOT00000086669 |
LOC100911215
|
T-cell surface glycoprotein CD5-like |
| chr3_-_3691972 | 0.06 |
ENSRNOT00000061735
|
Qsox2
|
quiescin sulfhydryl oxidase 2 |
| chr11_+_47188495 | 0.06 |
ENSRNOT00000002188
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr20_+_13827660 | 0.06 |
ENSRNOT00000093131
|
Ddt
|
D-dopachrome tautomerase |
| chr1_-_7480825 | 0.06 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr15_-_27815261 | 0.06 |
ENSRNOT00000032992
|
Klhl33
|
kelch-like family member 33 |
| chr7_+_117951154 | 0.06 |
ENSRNOT00000060181
|
Znf7
|
zinc finger protein 7 |
| chr3_+_172195844 | 0.06 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr1_+_264741911 | 0.06 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr12_-_29952196 | 0.06 |
ENSRNOT00000086175
|
Tmem248
|
transmembrane protein 248 |
| chr5_+_103479767 | 0.06 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chr15_+_38341089 | 0.06 |
ENSRNOT00000015367
|
Fgf9
|
fibroblast growth factor 9 |
| chr11_-_70211701 | 0.06 |
ENSRNOT00000076114
|
Muc13
|
mucin 13, cell surface associated |
| chr3_+_153197644 | 0.05 |
ENSRNOT00000008386
|
Tldc2
|
TBC/LysM-associated domain containing 2 |
| chr4_-_30556814 | 0.05 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr8_+_116332796 | 0.05 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr2_-_187706300 | 0.05 |
ENSRNOT00000092349
ENSRNOT00000026414 |
Tmem79
|
transmembrane protein 79 |
| chrX_+_54062935 | 0.05 |
ENSRNOT00000004854
|
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr7_-_98960736 | 0.05 |
ENSRNOT00000011984
ENSRNOT00000080155 |
Mtss1
|
MTSS1, I-BAR domain containing |
| chr16_-_74122889 | 0.05 |
ENSRNOT00000025763
|
Plat
|
plasminogen activator, tissue type |
| chr3_-_5975734 | 0.05 |
ENSRNOT00000081376
|
Vav2
|
vav guanine nucleotide exchange factor 2 |
| chr13_+_25778317 | 0.05 |
ENSRNOT00000021129
|
Tnfrsf11a
|
TNF receptor superfamily member 11A |
| chr10_+_14547172 | 0.05 |
ENSRNOT00000092043
|
Unkl
|
unkempt family like zinc finger |
| chr20_-_3819200 | 0.05 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr1_+_101884019 | 0.05 |
ENSRNOT00000028650
|
Tmem143
|
transmembrane protein 143 |
| chr2_+_196334626 | 0.05 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chrX_-_84768463 | 0.05 |
ENSRNOT00000088570
|
Chm
|
CHM, Rab escort protein 1 |
| chr4_+_57204944 | 0.05 |
ENSRNOT00000085027
|
Strip2
|
striatin interacting protein 2 |
| chr5_+_137257287 | 0.05 |
ENSRNOT00000037160
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr18_-_410098 | 0.05 |
ENSRNOT00000084138
|
LOC102546764
|
cx9C motif-containing protein 4-like |
| chr3_-_3660006 | 0.05 |
ENSRNOT00000045587
|
Lhx3
|
LIM homeobox 3 |
| chr1_-_204805738 | 0.05 |
ENSRNOT00000086830
|
Fam53b
|
family with sequence similarity 53, member B |
| chr9_+_98073038 | 0.05 |
ENSRNOT00000026795
|
Mlph
|
melanophilin |
| chr8_-_108109801 | 0.05 |
ENSRNOT00000034277
|
Sox14
|
SRY box 14 |
| chr14_-_86047162 | 0.05 |
ENSRNOT00000018227
|
Pgam2
|
phosphoglycerate mutase 2 |
| chr7_-_144960527 | 0.05 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr10_+_90731148 | 0.05 |
ENSRNOT00000093604
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr5_-_107841175 | 0.05 |
ENSRNOT00000079251
|
Cdkn2a
|
cyclin-dependent kinase inhibitor 2A |
| chrX_+_15610230 | 0.05 |
ENSRNOT00000012880
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr3_+_80075991 | 0.04 |
ENSRNOT00000080266
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr1_+_61733805 | 0.04 |
ENSRNOT00000086029
|
LOC682206
|
similar to Zinc finger protein 208 |
| chr13_+_82496022 | 0.04 |
ENSRNOT00000080759
|
F5
|
coagulation factor V |
| chr1_-_144601327 | 0.04 |
ENSRNOT00000029244
ENSRNOT00000078144 |
Saxo2
|
stabilizer of axonemal microtubules 2 |
| chr16_-_82099991 | 0.04 |
ENSRNOT00000089447
|
Atp11a
|
ATPase phospholipid transporting 11A |
| chrX_+_6273733 | 0.04 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr10_+_13230158 | 0.04 |
ENSRNOT00000075648
|
Csap1
|
common salivary protein 1 |
| chr1_+_86973745 | 0.04 |
ENSRNOT00000078156
|
Rinl
|
Ras and Rab interactor-like |
| chr13_-_48400632 | 0.04 |
ENSRNOT00000074204
|
Avpr1b
|
arginine vasopressin receptor 1B |
| chr10_+_63662986 | 0.04 |
ENSRNOT00000088722
|
Scarf1
|
scavenger receptor class F, member 1 |
| chr7_-_72328128 | 0.04 |
ENSRNOT00000008227
|
Tspyl5
|
TSPY-like 5 |
| chr20_-_4530126 | 0.04 |
ENSRNOT00000000481
|
Skiv2l
|
Ski2 like RNA helicase |
| chr1_+_220806473 | 0.04 |
ENSRNOT00000027869
|
Bles03
|
basophilic leukemia expressed protein BLES03 |
| chr15_-_33218456 | 0.04 |
ENSRNOT00000017753
|
Ajuba
|
ajuba LIM protein |
| chr16_-_82100222 | 0.04 |
ENSRNOT00000023163
|
Atp11a
|
ATPase phospholipid transporting 11A |
| chr10_+_86340940 | 0.04 |
ENSRNOT00000073486
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr1_-_98493978 | 0.04 |
ENSRNOT00000023942
|
Nkg7
|
natural killer cell granule protein 7 |
| chrX_+_136476823 | 0.04 |
ENSRNOT00000093183
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr6_+_56846789 | 0.04 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr7_-_23953002 | 0.04 |
ENSRNOT00000047587
|
RGD1563124
|
similar to 40S ribosomal protein S20 |
| chr3_-_168033457 | 0.04 |
ENSRNOT00000055111
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr7_+_118490560 | 0.04 |
ENSRNOT00000064873
|
LOC108348145
|
zinc finger protein 7-like |
| chr10_+_105568091 | 0.04 |
ENSRNOT00000015221
|
Aanat
|
aralkylamine N-acetyltransferase |
| chr1_+_101884276 | 0.04 |
ENSRNOT00000082917
|
Tmem143
|
transmembrane protein 143 |
| chr10_-_12916784 | 0.04 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr15_+_23619123 | 0.04 |
ENSRNOT00000013912
|
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr7_-_114848414 | 0.04 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr2_+_86914989 | 0.04 |
ENSRNOT00000085164
ENSRNOT00000081966 |
LOC100360380
|
zinc finger protein 457-like |
| chr4_+_153876149 | 0.04 |
ENSRNOT00000018083
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr15_+_34520142 | 0.04 |
ENSRNOT00000074659
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr5_+_126812187 | 0.04 |
ENSRNOT00000057372
|
Ldlrad1
|
low density lipoprotein receptor class A domain containing 1 |
| chr10_-_18443934 | 0.04 |
ENSRNOT00000059895
ENSRNOT00000080021 |
Ranbp17
|
RAN binding protein 17 |
| chr14_+_70164650 | 0.04 |
ENSRNOT00000004385
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr1_-_213987053 | 0.04 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chr10_+_90731865 | 0.03 |
ENSRNOT00000064429
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr1_+_177249226 | 0.03 |
ENSRNOT00000021671
|
Parva
|
parvin, alpha |
| chr7_-_123101851 | 0.03 |
ENSRNOT00000090984
ENSRNOT00000005837 |
Phf5a
|
PHD finger protein 5A |
| chr13_-_73056765 | 0.03 |
ENSRNOT00000000049
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr18_-_29340403 | 0.03 |
ENSRNOT00000025157
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr3_+_155269347 | 0.03 |
ENSRNOT00000021308
|
Fam83d
|
family with sequence similarity 83, member D |
| chr12_-_2438817 | 0.03 |
ENSRNOT00000037059
|
Ccl25
|
C-C motif chemokine ligand 25 |
| chr8_+_39848448 | 0.03 |
ENSRNOT00000012248
|
Hepacam
|
hepatocyte cell adhesion molecule |
| chr1_-_154170409 | 0.03 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr13_-_98530724 | 0.03 |
ENSRNOT00000040203
|
Psen2
|
presenilin 2 |
| chr9_+_38297322 | 0.03 |
ENSRNOT00000078157
ENSRNOT00000088824 |
Bend6
|
BEN domain containing 6 |
| chr4_+_77211692 | 0.03 |
ENSRNOT00000007620
|
Cul1
|
cullin 1 |
| chr8_+_28075551 | 0.03 |
ENSRNOT00000012078
|
Ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr1_-_91042230 | 0.03 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr11_+_61531416 | 0.03 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr5_-_40867023 | 0.03 |
ENSRNOT00000011576
|
Manea
|
mannosidase, endo-alpha |
| chr8_-_130617833 | 0.03 |
ENSRNOT00000026363
|
Pomgnt2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr4_-_168382837 | 0.03 |
ENSRNOT00000029510
|
Mansc1
|
MANSC domain containing 1 |
| chr17_+_42302540 | 0.03 |
ENSRNOT00000025389
|
Gmnn
|
geminin, DNA replication inhibitor |
| chr16_+_20109200 | 0.03 |
ENSRNOT00000079784
|
Jak3
|
Janus kinase 3 |
| chr4_+_78140483 | 0.03 |
ENSRNOT00000048576
|
Zfp862
|
zinc finger protein 862 |
| chr3_+_1452644 | 0.03 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr3_+_12262822 | 0.03 |
ENSRNOT00000022585
|
Angptl2
|
angiopoietin-like 2 |
| chr13_-_89745835 | 0.03 |
ENSRNOT00000005352
|
Klhdc9
|
kelch domain containing 9 |
| chr2_+_3662763 | 0.03 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr13_+_48287873 | 0.03 |
ENSRNOT00000068223
|
Fam72a
|
family with sequence similarity 72, member A |
| chr20_-_7482747 | 0.03 |
ENSRNOT00000038195
|
Taf11
|
TATA-box binding protein associated factor 11 |
| chr16_-_71040847 | 0.03 |
ENSRNOT00000020606
|
Star
|
steroidogenic acute regulatory protein |
| chr1_+_31264755 | 0.03 |
ENSRNOT00000028988
|
LOC679739
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr7_+_141973553 | 0.03 |
ENSRNOT00000052075
|
Mettl7a
|
methyltransferase like 7A |
| chr1_+_221236773 | 0.03 |
ENSRNOT00000051979
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr10_+_11146359 | 0.03 |
ENSRNOT00000006314
|
Pam16
|
presequence translocase associated motor 16 homolog |
| chr12_+_16988136 | 0.03 |
ENSRNOT00000078925
ENSRNOT00000036744 |
Micall2
|
MICAL-like 2 |
| chr14_+_9226125 | 0.03 |
ENSRNOT00000088047
|
Wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr9_-_30321323 | 0.03 |
ENSRNOT00000018234
|
Fam135a
|
family with sequence similarity 135, member A |
| chr4_+_88832178 | 0.03 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr17_+_90670872 | 0.03 |
ENSRNOT00000081530
|
Gpr137b
|
G protein-coupled receptor 137B |
| chr1_-_101651668 | 0.03 |
ENSRNOT00000028519
|
Fut2
|
fucosyltransferase 2 |
| chr3_+_81287242 | 0.03 |
ENSRNOT00000086530
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr2_+_56426367 | 0.03 |
ENSRNOT00000016036
|
Lifr
|
leukemia inhibitory factor receptor alpha |
| chr9_+_10216205 | 0.03 |
ENSRNOT00000084312
ENSRNOT00000073025 |
Rfx2
|
regulatory factor X2 |
| chr14_+_46001849 | 0.03 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr4_+_140886545 | 0.03 |
ENSRNOT00000088273
|
Edem1
|
ER degradation enhancing alpha-mannosidase like protein 1 |
| chr7_-_139734568 | 0.03 |
ENSRNOT00000079377
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr19_+_44164935 | 0.03 |
ENSRNOT00000048998
|
Gabarapl2
|
GABA type A receptor associated protein like 2 |
| chr1_-_156296161 | 0.03 |
ENSRNOT00000025653
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr5_+_165415136 | 0.03 |
ENSRNOT00000016317
ENSRNOT00000079407 |
Masp2
|
mannan-binding lectin serine peptidase 2 |
| chr13_+_110511668 | 0.03 |
ENSRNOT00000006235
|
Nek2
|
NIMA-related kinase 2 |
| chr12_-_2170504 | 0.03 |
ENSRNOT00000001309
|
Xab2
|
XPA binding protein 2 |
| chr19_-_55389256 | 0.03 |
ENSRNOT00000064180
|
Aprt
|
adenine phosphoribosyl transferase |
| chr7_+_139685573 | 0.03 |
ENSRNOT00000088376
|
Pfkm
|
phosphofructokinase, muscle |
| chr8_+_117836701 | 0.03 |
ENSRNOT00000043345
|
Plxnb1
|
plexin B1 |
| chr8_-_50231357 | 0.03 |
ENSRNOT00000081737
|
Tagln
|
transgelin |
| chr11_+_28692708 | 0.03 |
ENSRNOT00000002135
|
Krtap13-1
|
keratin associated protein 13-1 |
| chr19_+_14489983 | 0.03 |
ENSRNOT00000075435
|
Zfp14
|
ZFP14 zinc finger protein |
| chr3_+_79728796 | 0.03 |
ENSRNOT00000068124
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr7_-_70926903 | 0.03 |
ENSRNOT00000031005
|
Lrp1
|
LDL receptor related protein 1 |
| chr4_+_7377563 | 0.03 |
ENSRNOT00000084826
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr7_-_121302740 | 0.03 |
ENSRNOT00000067032
|
Rpl3
|
ribosomal protein L3 |
| chr1_+_100393303 | 0.03 |
ENSRNOT00000026251
|
Syt3
|
synaptotagmin 3 |
| chr6_-_109205004 | 0.03 |
ENSRNOT00000010512
|
Tmed10
|
transmembrane p24 trafficking protein 10 |
| chr10_-_57618527 | 0.02 |
ENSRNOT00000037517
|
C1qbp
|
complement C1q binding protein |
| chr5_+_137257637 | 0.02 |
ENSRNOT00000093001
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr5_-_156781291 | 0.02 |
ENSRNOT00000075128
|
Cda
|
cytidine deaminase |
| chr2_-_149088787 | 0.02 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr13_+_49092306 | 0.02 |
ENSRNOT00000000039
|
Nuak2
|
NUAK family kinase 2 |
| chr10_-_77512032 | 0.02 |
ENSRNOT00000003295
|
Pctp
|
phosphatidylcholine transfer protein |
| chr7_-_60825242 | 0.02 |
ENSRNOT00000009203
|
Nup107
|
nucleoporin 107 |
| chr1_+_221612584 | 0.02 |
ENSRNOT00000090100
|
Atg2a
|
autophagy related 2A |
| chr14_+_108412152 | 0.02 |
ENSRNOT00000083707
|
Pus10
|
pseudouridylate synthase 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2c2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.1 | 0.2 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0034443 | plasma lipoprotein particle oxidation(GO:0034441) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0070268 | cuticle development(GO:0042335) cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0019483 | beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.1 | GO:0071847 | circadian temperature homeostasis(GO:0060086) TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.0 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.0 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.0 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.0 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.1 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.0 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 0.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |