Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
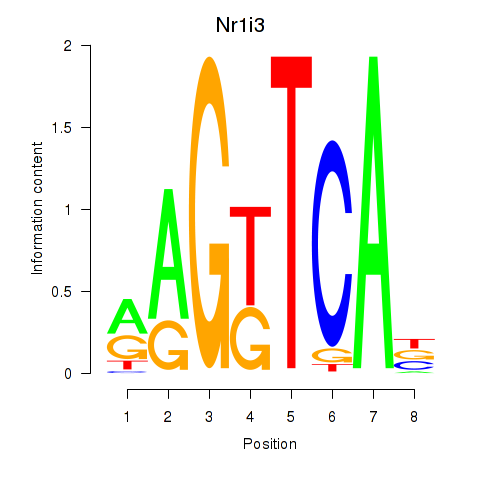
Results for Nr1i3
Z-value: 0.58
Transcription factors associated with Nr1i3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1i3
|
ENSRNOG00000003260 | nuclear receptor subfamily 1, group I, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1i3 | rn6_v1_chr13_+_89586283_89586336 | 0.22 | 7.2e-01 | Click! |
Activity profile of Nr1i3 motif
Sorted Z-values of Nr1i3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_40227297 | 0.19 |
ENSRNOT00000030600
|
Fam109a
|
family with sequence similarity 109, member A |
| chr8_-_132345708 | 0.18 |
ENSRNOT00000006300
|
Tmem158
|
transmembrane protein 158 |
| chr3_+_152571121 | 0.18 |
ENSRNOT00000087289
ENSRNOT00000080543 ENSRNOT00000083476 |
Epb41l1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr17_-_61332391 | 0.18 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr2_-_259150479 | 0.17 |
ENSRNOT00000085892
|
AABR07013843.1
|
|
| chr16_+_73755539 | 0.17 |
ENSRNOT00000088219
|
LOC100910418
|
tissue-type plasminogen activator-like |
| chrX_-_106607352 | 0.17 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr7_+_121311024 | 0.16 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr13_-_70667658 | 0.16 |
ENSRNOT00000092521
|
Lamc1
|
laminin subunit gamma 1 |
| chr6_-_135939534 | 0.15 |
ENSRNOT00000052237
|
Diras3
|
DIRAS family GTPase 3 |
| chr1_-_201906286 | 0.15 |
ENSRNOT00000064511
ENSRNOT00000036625 |
RGD1560958
|
similar to RIKEN cDNA 1700063I17 |
| chr2_-_198412350 | 0.14 |
ENSRNOT00000040210
|
LOC100912489
|
histone H4-like |
| chr12_-_22245100 | 0.14 |
ENSRNOT00000001912
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr4_+_78263866 | 0.14 |
ENSRNOT00000033807
|
AI854703
|
expressed sequence AI854703 |
| chr3_+_147609095 | 0.14 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr1_-_80744831 | 0.14 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr5_+_74874306 | 0.14 |
ENSRNOT00000076918
|
Akap2
|
A-kinase anchoring protein 2 |
| chr5_-_152473868 | 0.14 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr4_-_108717309 | 0.14 |
ENSRNOT00000085062
|
AABR07061178.1
|
|
| chr8_+_116776494 | 0.14 |
ENSRNOT00000050702
|
Cdhr4
|
cadherin-related family member 4 |
| chr20_+_6351458 | 0.13 |
ENSRNOT00000091731
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr10_-_85435016 | 0.13 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr2_-_225107283 | 0.13 |
ENSRNOT00000055711
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr8_+_118662335 | 0.13 |
ENSRNOT00000029755
|
Ngp
|
neutrophilic granule protein |
| chr4_-_100783750 | 0.13 |
ENSRNOT00000078956
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chrX_+_15113878 | 0.12 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr6_+_26390689 | 0.12 |
ENSRNOT00000079762
ENSRNOT00000091528 |
Ift172
|
intraflagellar transport 172 |
| chr1_+_78876205 | 0.12 |
ENSRNOT00000022610
|
Pnmal2
|
paraneoplastic Ma antigen family-like 2 |
| chr2_+_235596907 | 0.12 |
ENSRNOT00000071463
ENSRNOT00000075728 |
Col25a1
|
collagen type XXV alpha 1 chain |
| chr4_+_119815139 | 0.12 |
ENSRNOT00000083402
ENSRNOT00000016137 |
Hmces
|
5-hydroxymethylcytosine (hmC) binding, ES cell-specific |
| chr1_+_90948976 | 0.12 |
ENSRNOT00000056877
|
LOC108348111
|
succinate dehydrogenase assembly factor 1, mitochondrial |
| chr14_-_114649173 | 0.12 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr3_-_153876938 | 0.12 |
ENSRNOT00000087907
|
LOC100911769
|
band 4.1-like protein 1-like |
| chr13_+_110920830 | 0.12 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr12_-_30304036 | 0.12 |
ENSRNOT00000001218
|
Nupr2
|
nuclear protein 2, transcriptional regulator |
| chr19_+_740028 | 0.12 |
ENSRNOT00000076604
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr4_+_100209951 | 0.12 |
ENSRNOT00000015807
|
LOC691113
|
hypothetical protein LOC691113 |
| chr11_+_89008008 | 0.12 |
ENSRNOT00000074586
|
Cebpd
|
CCAAT/enhancer binding protein delta |
| chr12_+_37878653 | 0.11 |
ENSRNOT00000091084
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr13_-_88642011 | 0.11 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr1_-_215284548 | 0.11 |
ENSRNOT00000090375
|
Cttn
|
cortactin |
| chr1_-_204275803 | 0.11 |
ENSRNOT00000066711
|
Chst15
|
carbohydrate sulfotransferase 15 |
| chr10_-_103848035 | 0.11 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr12_-_2610917 | 0.11 |
ENSRNOT00000034124
|
Evi5l
|
ecotropic viral integration site 5-like |
| chr7_+_127081978 | 0.11 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr6_-_41870046 | 0.11 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr3_-_13525983 | 0.11 |
ENSRNOT00000082036
|
Pbx3
|
PBX homeobox 3 |
| chr7_-_44121130 | 0.11 |
ENSRNOT00000005706
|
Nts
|
neurotensin |
| chr12_-_2555164 | 0.11 |
ENSRNOT00000084460
ENSRNOT00000061821 |
Map2k7
|
mitogen activated protein kinase kinase 7 |
| chr7_-_12275609 | 0.11 |
ENSRNOT00000086061
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chr1_+_213583606 | 0.11 |
ENSRNOT00000088899
|
AC109844.1
|
|
| chr4_+_64088900 | 0.11 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr11_+_61531416 | 0.11 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr6_+_132219088 | 0.11 |
ENSRNOT00000034883
|
Hhipl1
|
HHIP-like 1 |
| chr1_-_142724511 | 0.10 |
ENSRNOT00000014639
|
Nmb
|
neuromedin B |
| chr6_-_104631355 | 0.10 |
ENSRNOT00000007825
|
Slc10a1
|
solute carrier family 10 member 1 |
| chr16_+_20578131 | 0.10 |
ENSRNOT00000026713
|
Ssbp4
|
single stranded DNA binding protein 4 |
| chr12_-_47987255 | 0.10 |
ENSRNOT00000074000
|
Ube3b
|
ubiquitin protein ligase E3B |
| chr20_+_3149114 | 0.10 |
ENSRNOT00000084770
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr1_+_199449973 | 0.10 |
ENSRNOT00000029994
|
Trim72
|
tripartite motif containing 72 |
| chr5_-_17061837 | 0.10 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr4_+_199916 | 0.10 |
ENSRNOT00000009317
|
Htr5a
|
5-hydroxytryptamine receptor 5A |
| chr6_+_76677213 | 0.10 |
ENSRNOT00000076760
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr3_-_120306551 | 0.10 |
ENSRNOT00000021082
|
Mall
|
mal, T-cell differentiation protein-like |
| chr17_-_14058642 | 0.10 |
ENSRNOT00000019489
|
Nxnl2
|
nucleoredoxin-like 2 |
| chr20_+_4993560 | 0.10 |
ENSRNOT00000081628
ENSRNOT00000087861 ENSRNOT00000001160 |
Vars
|
valyl-tRNA synthetase |
| chr12_-_35979193 | 0.10 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr9_+_10471742 | 0.10 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr14_+_86652365 | 0.10 |
ENSRNOT00000077428
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr17_+_76079720 | 0.10 |
ENSRNOT00000073933
|
Proser2
|
proline and serine rich 2 |
| chr1_+_80279706 | 0.10 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr7_+_20462081 | 0.10 |
ENSRNOT00000088383
|
AABR07056131.1
|
|
| chr18_-_56325095 | 0.10 |
ENSRNOT00000025209
|
Slc6a7
|
solute carrier family 6 member 7 |
| chr10_-_105368242 | 0.10 |
ENSRNOT00000075293
ENSRNOT00000072230 |
Rnf157
|
ring finger protein 157 |
| chr12_+_32103198 | 0.09 |
ENSRNOT00000085464
|
Tmem132d
|
transmembrane protein 132D |
| chr4_+_170149029 | 0.09 |
ENSRNOT00000073287
|
LOC103690002
|
histone H2A.J |
| chr1_-_200548317 | 0.09 |
ENSRNOT00000088610
|
AABR07005806.1
|
|
| chr19_-_36239712 | 0.09 |
ENSRNOT00000071962
|
AABR07043711.2
|
|
| chr10_+_103737162 | 0.09 |
ENSRNOT00000055037
|
Tmem104
|
transmembrane protein 104 |
| chr11_-_25350974 | 0.09 |
ENSRNOT00000002187
|
Adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr9_-_10659357 | 0.09 |
ENSRNOT00000091450
|
Kdm4b
|
lysine demethylase 4B |
| chr8_-_71728654 | 0.09 |
ENSRNOT00000031207
ENSRNOT00000091751 |
Snx22
|
sorting nexin 22 |
| chr16_-_70705128 | 0.09 |
ENSRNOT00000071258
|
Rnf170
|
ring finger protein 170 |
| chr17_+_76002275 | 0.09 |
ENSRNOT00000092665
ENSRNOT00000086701 |
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr8_+_49441106 | 0.09 |
ENSRNOT00000030152
|
Scn4b
|
sodium voltage-gated channel beta subunit 4 |
| chr3_+_145764932 | 0.09 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chr19_+_24800072 | 0.09 |
ENSRNOT00000005470
|
Ptger1
|
prostaglandin E receptor 1 |
| chr6_+_10348308 | 0.09 |
ENSRNOT00000034991
|
Epas1
|
endothelial PAS domain protein 1 |
| chr3_+_103773459 | 0.09 |
ENSRNOT00000079727
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr7_-_98735192 | 0.09 |
ENSRNOT00000051430
|
AABR07058017.1
|
|
| chr14_+_1355004 | 0.09 |
ENSRNOT00000043131
|
AABR07014114.1
|
|
| chr11_+_83855753 | 0.09 |
ENSRNOT00000030686
|
RGD1563956
|
similar to 60S ribosomal protein L12 |
| chr1_+_242572533 | 0.09 |
ENSRNOT00000035123
|
Tmem252
|
transmembrane protein 252 |
| chr9_-_15628881 | 0.09 |
ENSRNOT00000077318
ENSRNOT00000020950 |
Guca1b
|
guanylate cyclase activator 1B |
| chr4_+_40161285 | 0.09 |
ENSRNOT00000050722
|
LOC500035
|
hypothetical protein LOC500035 |
| chr10_+_47019326 | 0.09 |
ENSRNOT00000006984
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chrX_+_111735820 | 0.09 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr2_+_260997497 | 0.09 |
ENSRNOT00000012638
|
Erich3
|
glutamate-rich 3 |
| chr10_+_63829807 | 0.08 |
ENSRNOT00000006407
|
Crk
|
CRK proto-oncogene, adaptor protein |
| chr1_+_166125474 | 0.08 |
ENSRNOT00000091822
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr7_+_70283710 | 0.08 |
ENSRNOT00000075371
|
Ctdsp2
|
CTD small phosphatase 2 |
| chr14_-_114583122 | 0.08 |
ENSRNOT00000084595
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr1_+_214927172 | 0.08 |
ENSRNOT00000027134
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr15_-_70399924 | 0.08 |
ENSRNOT00000087940
|
Diaph3
|
diaphanous-related formin 3 |
| chr8_+_49418965 | 0.08 |
ENSRNOT00000021819
|
Scn2b
|
sodium voltage-gated channel beta subunit 2 |
| chr7_+_130474508 | 0.08 |
ENSRNOT00000085191
|
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr10_+_57218087 | 0.08 |
ENSRNOT00000089853
|
Mink1
|
misshapen-like kinase 1 |
| chr9_-_28732919 | 0.08 |
ENSRNOT00000083915
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr12_+_25119355 | 0.08 |
ENSRNOT00000034629
|
Lat2
|
linker for activation of T cells family, member 2 |
| chr7_+_130498199 | 0.08 |
ENSRNOT00000092684
ENSRNOT00000092431 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr10_-_90994437 | 0.08 |
ENSRNOT00000093167
|
Gfap
|
glial fibrillary acidic protein |
| chr5_-_105579959 | 0.08 |
ENSRNOT00000010827
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr1_+_203971152 | 0.08 |
ENSRNOT00000075540
|
Gpr26
|
G protein-coupled receptor 26 |
| chr8_+_118378059 | 0.08 |
ENSRNOT00000043247
|
AABR07071482.1
|
|
| chr1_+_27924825 | 0.08 |
ENSRNOT00000087830
|
AABR07000902.1
|
|
| chr10_-_63952726 | 0.08 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr5_+_16463597 | 0.08 |
ENSRNOT00000091550
|
AABR07047036.1
|
|
| chr14_+_79205466 | 0.08 |
ENSRNOT00000085534
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chr2_-_62634785 | 0.08 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr19_+_755460 | 0.08 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr16_+_19051965 | 0.08 |
ENSRNOT00000016399
|
Slc35e1
|
solute carrier family 35, member E1 |
| chr20_-_14620019 | 0.08 |
ENSRNOT00000001779
|
Gnaz
|
G protein subunit alpha z |
| chr1_+_24900858 | 0.08 |
ENSRNOT00000075616
|
LOC501467
|
similar to spermatogenesis associated glutamate (E)-rich protein 4d |
| chr6_+_145770662 | 0.08 |
ENSRNOT00000088969
|
Cdca7l
|
cell division cycle associated 7 like |
| chr9_-_10471009 | 0.07 |
ENSRNOT00000072868
|
Safb
|
scaffold attachment factor B |
| chr20_+_4824226 | 0.07 |
ENSRNOT00000001113
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr16_+_70081035 | 0.07 |
ENSRNOT00000016098
|
LOC100912059
|
ubiquitin carboxyl-terminal hydrolase 12-like |
| chr2_+_206064179 | 0.07 |
ENSRNOT00000025953
|
Syt6
|
synaptotagmin 6 |
| chr14_+_42015347 | 0.07 |
ENSRNOT00000044017
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr8_+_116332796 | 0.07 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr2_-_200003443 | 0.07 |
ENSRNOT00000024900
ENSRNOT00000088041 |
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr1_+_199495298 | 0.07 |
ENSRNOT00000086003
ENSRNOT00000026748 |
Itgad
|
integrin subunit alpha D |
| chr17_-_43815183 | 0.07 |
ENSRNOT00000073188
|
Hist1h2ail1
|
histone cluster 1, H2ai-like1 |
| chr4_-_155563249 | 0.07 |
ENSRNOT00000011298
|
Slc2a3
|
solute carrier family 2 member 3 |
| chr8_-_73122740 | 0.07 |
ENSRNOT00000012026
|
Tln2
|
talin 2 |
| chr12_-_30770791 | 0.07 |
ENSRNOT00000093734
|
Sfswap
|
splicing factor SWAP homolog |
| chr6_+_137164535 | 0.07 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr19_-_54648227 | 0.07 |
ENSRNOT00000025424
|
Klhdc4
|
kelch domain containing 4 |
| chr5_-_137372524 | 0.07 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr7_+_12471824 | 0.07 |
ENSRNOT00000068197
|
Sbno2
|
strawberry notch homolog 2 |
| chrX_+_158351156 | 0.07 |
ENSRNOT00000080538
|
LOC100909732
|
protein FAM122B-like |
| chr2_-_115891097 | 0.07 |
ENSRNOT00000013191
|
Skil
|
SKI-like proto-oncogene |
| chr4_+_117743710 | 0.07 |
ENSRNOT00000021491
|
Add2
|
adducin 2 |
| chr4_-_179477070 | 0.07 |
ENSRNOT00000030680
|
Casc1
|
cancer susceptibility candidate 1 |
| chr11_+_32440237 | 0.07 |
ENSRNOT00000040844
|
Kcne2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chr1_+_84411726 | 0.07 |
ENSRNOT00000025303
|
Akt2
|
AKT serine/threonine kinase 2 |
| chr11_-_30051103 | 0.07 |
ENSRNOT00000046486
|
Tiam1
|
T-cell lymphoma invasion and metastasis 1 |
| chr8_-_118378460 | 0.07 |
ENSRNOT00000047247
|
RGD1563705
|
similar to ribosomal protein S23 |
| chr10_+_56381813 | 0.07 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr10_-_5196892 | 0.07 |
ENSRNOT00000083982
|
Clec16a
|
C-type lectin domain family 16, member A |
| chr2_+_187990242 | 0.07 |
ENSRNOT00000092819
|
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr8_+_22047697 | 0.07 |
ENSRNOT00000067741
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr2_+_45668969 | 0.07 |
ENSRNOT00000014761
ENSRNOT00000071353 |
Arl15
|
ADP-ribosylation factor like GTPase 15 |
| chr10_+_47018974 | 0.07 |
ENSRNOT00000079375
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr1_+_72882806 | 0.07 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr6_+_146784915 | 0.07 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr13_+_51795867 | 0.07 |
ENSRNOT00000006747
|
Ube2t
|
ubiquitin-conjugating enzyme E2T |
| chr20_+_3823596 | 0.07 |
ENSRNOT00000087670
|
Rxrb
|
retinoid X receptor beta |
| chr3_+_162346490 | 0.07 |
ENSRNOT00000087530
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chrX_+_15114892 | 0.07 |
ENSRNOT00000078168
|
Wdr13
|
WD repeat domain 13 |
| chr10_-_14299167 | 0.07 |
ENSRNOT00000042066
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr2_-_77632628 | 0.07 |
ENSRNOT00000073915
|
Basp1
|
brain abundant, membrane attached signal protein 1 |
| chr3_+_71114100 | 0.07 |
ENSRNOT00000088549
ENSRNOT00000006961 |
Itgav
|
integrin subunit alpha V |
| chr4_-_176679815 | 0.07 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr9_-_10462491 | 0.07 |
ENSRNOT00000081770
|
Safb
|
scaffold attachment factor B |
| chr3_+_151126591 | 0.06 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr4_+_145427367 | 0.06 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chr14_-_5006594 | 0.06 |
ENSRNOT00000076571
|
Zfp326
|
zinc finger protein 326 |
| chr9_+_98190829 | 0.06 |
ENSRNOT00000050191
|
Lrrfip1
|
LRR binding FLII interacting protein 1 |
| chr2_-_257474733 | 0.06 |
ENSRNOT00000016967
ENSRNOT00000066780 |
Nexn
|
nexilin (F actin binding protein) |
| chr7_-_26690323 | 0.06 |
ENSRNOT00000087536
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr10_+_46092754 | 0.06 |
ENSRNOT00000080666
|
Mprip
|
myosin phosphatase Rho interacting protein |
| chr19_+_43882249 | 0.06 |
ENSRNOT00000025761
|
Zfp1
|
zinc finger protein 1 |
| chr12_-_37910439 | 0.06 |
ENSRNOT00000001433
|
Ogfod2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr17_+_44758460 | 0.06 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chr19_+_52664322 | 0.06 |
ENSRNOT00000082754
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr1_-_72366455 | 0.06 |
ENSRNOT00000031786
|
LOC100911196
|
zinc finger protein 865-like |
| chr11_+_69484293 | 0.06 |
ENSRNOT00000049292
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr6_+_106039991 | 0.06 |
ENSRNOT00000088917
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr14_+_83724933 | 0.06 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr9_+_10773901 | 0.06 |
ENSRNOT00000070978
|
Plin3
|
perilipin 3 |
| chr7_-_125497691 | 0.06 |
ENSRNOT00000049445
|
AABR07058578.1
|
|
| chr10_+_40553180 | 0.06 |
ENSRNOT00000087763
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr5_+_124476168 | 0.06 |
ENSRNOT00000077754
|
RGD1564074
|
similar to novel protein |
| chr3_+_8701855 | 0.06 |
ENSRNOT00000021431
|
Tbc1d13
|
TBC1 domain family, member 13 |
| chr11_+_64472072 | 0.06 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr3_-_155192806 | 0.06 |
ENSRNOT00000051343
|
RGD1563145
|
similar to 60S ribosomal protein L13 |
| chr1_-_16203838 | 0.06 |
ENSRNOT00000018556
ENSRNOT00000078586 |
Pde7b
|
phosphodiesterase 7B |
| chr3_+_171832500 | 0.06 |
ENSRNOT00000007554
|
Vapb
|
VAMP associated protein B and C |
| chr10_+_90731865 | 0.06 |
ENSRNOT00000064429
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr4_-_77706994 | 0.06 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr12_-_48857724 | 0.06 |
ENSRNOT00000080783
|
Wscd2
|
WSC domain containing 2 |
| chr5_+_172273459 | 0.06 |
ENSRNOT00000017957
|
Mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr3_-_6626284 | 0.06 |
ENSRNOT00000012494
|
Fcnb
|
ficolin B |
| chr7_-_15073052 | 0.06 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr8_-_55276070 | 0.06 |
ENSRNOT00000041555
|
LOC108351703
|
60S ribosomal protein L27a-like |
| chr10_+_109893700 | 0.06 |
ENSRNOT00000082551
|
Lrrc45
|
leucine rich repeat containing 45 |
| chr19_-_11302938 | 0.06 |
ENSRNOT00000038212
|
AC128848.1
|
|
| chr10_-_40375605 | 0.06 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr10_-_50402616 | 0.06 |
ENSRNOT00000004546
|
Hs3st3b1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1i3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.2 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:1904054 | regulation of cholangiocyte proliferation(GO:1904054) positive regulation of cholangiocyte proliferation(GO:1904056) |
| 0.0 | 0.1 | GO:0031439 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 0.0 | 0.1 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0001997 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.0 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:1990859 | response to endothelin(GO:1990839) cellular response to endothelin(GO:1990859) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.1 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.0 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.0 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.0 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.0 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.0 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) |
| 0.0 | 0.0 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.0 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.0 | GO:1904373 | response to kainic acid(GO:1904373) |
| 0.0 | 0.0 | GO:1903373 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.0 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:2001137 | actin filament uncapping(GO:0051695) positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0061525 | hindgut development(GO:0061525) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.0 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.1 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.0 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.0 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.0 | GO:0031309 | intrinsic component of nuclear outer membrane(GO:0031308) integral component of nuclear outer membrane(GO:0031309) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.2 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0033222 | xylose binding(GO:0033222) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0038132 | C-X3-C chemokine binding(GO:0019960) neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.0 | GO:0000250 | lanosterol synthase activity(GO:0000250) oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.0 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.0 | GO:0047522 | 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |