Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
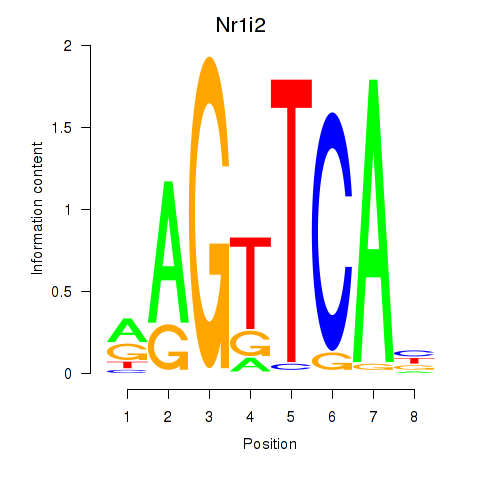
Results for Nr1i2
Z-value: 0.50
Transcription factors associated with Nr1i2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1i2
|
ENSRNOG00000002906 | nuclear receptor subfamily 1, group I, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1i2 | rn6_v1_chr11_+_65022100_65022100 | -0.69 | 2.0e-01 | Click! |
Activity profile of Nr1i2 motif
Sorted Z-values of Nr1i2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_58771908 | 0.27 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chr8_-_84506328 | 0.21 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr9_-_14599594 | 0.19 |
ENSRNOT00000018138
|
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr8_+_26652121 | 0.18 |
ENSRNOT00000009342
|
Eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chrX_+_156873849 | 0.18 |
ENSRNOT00000085410
|
Arhgap4
|
Rho GTPase activating protein 4 |
| chr1_+_141767940 | 0.18 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr6_+_29518416 | 0.16 |
ENSRNOT00000078660
|
AABR07063346.1
|
|
| chr13_-_111948753 | 0.16 |
ENSRNOT00000048074
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr8_-_119889661 | 0.15 |
ENSRNOT00000011780
|
Stac
|
SH3 and cysteine rich domain |
| chr1_+_166125474 | 0.15 |
ENSRNOT00000091822
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr17_+_9236249 | 0.14 |
ENSRNOT00000015878
|
Tifab
|
TIFA inhibitor |
| chr13_-_86671515 | 0.14 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr20_+_28572242 | 0.14 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr5_-_147635789 | 0.13 |
ENSRNOT00000037106
|
Zbtb8b
|
zinc finger and BTB domain containing 8b |
| chr1_-_90520344 | 0.13 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr2_-_207300854 | 0.12 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr4_-_169999873 | 0.12 |
ENSRNOT00000011697
|
Grin2b
|
glutamate ionotropic receptor NMDA type subunit 2B |
| chr2_+_189714754 | 0.12 |
ENSRNOT00000086285
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr6_+_112203679 | 0.12 |
ENSRNOT00000031205
|
Nrxn3
|
neurexin 3 |
| chr7_-_2961873 | 0.12 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chr2_+_235596907 | 0.12 |
ENSRNOT00000071463
ENSRNOT00000075728 |
Col25a1
|
collagen type XXV alpha 1 chain |
| chr11_+_82848853 | 0.12 |
ENSRNOT00000073743
|
Thpo
|
thrombopoietin |
| chr2_-_247988462 | 0.12 |
ENSRNOT00000022387
|
Pdlim5
|
PDZ and LIM domain 5 |
| chrX_-_106607352 | 0.11 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chr10_-_36419926 | 0.11 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr1_-_167911961 | 0.11 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr3_+_58632476 | 0.11 |
ENSRNOT00000010630
|
Rapgef4
|
Rap guanine nucleotide exchange factor 4 |
| chr12_-_39396042 | 0.11 |
ENSRNOT00000001746
|
P2rx7
|
purinergic receptor P2X 7 |
| chr7_+_34533543 | 0.10 |
ENSRNOT00000007412
ENSRNOT00000084185 |
Ntn4
|
netrin 4 |
| chr4_-_155563249 | 0.10 |
ENSRNOT00000011298
|
Slc2a3
|
solute carrier family 2 member 3 |
| chr10_+_86795787 | 0.10 |
ENSRNOT00000029021
|
Wipf2
|
WAS/WASL interacting protein family, member 2 |
| chr10_-_64642292 | 0.10 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr4_-_27473150 | 0.10 |
ENSRNOT00000032505
|
Krit1
|
KRIT1, ankyrin repeat containing |
| chr2_+_234375315 | 0.10 |
ENSRNOT00000071270
|
LOC102549542
|
elongation of very long chain fatty acids protein 6-like |
| chr17_+_49417067 | 0.10 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr7_+_2529077 | 0.09 |
ENSRNOT00000079383
|
Baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr13_-_105140473 | 0.09 |
ENSRNOT00000087023
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr18_+_30913842 | 0.09 |
ENSRNOT00000026947
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3 |
| chr7_-_143966863 | 0.09 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr6_-_114476723 | 0.09 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chrX_-_70428364 | 0.09 |
ENSRNOT00000045907
|
P2ry4
|
pyrimidinergic receptor P2Y4 |
| chr2_-_231521052 | 0.09 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr5_-_136098013 | 0.09 |
ENSRNOT00000089166
|
RGD1563714
|
RGD1563714 |
| chr11_+_69484293 | 0.09 |
ENSRNOT00000049292
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr5_+_140712583 | 0.09 |
ENSRNOT00000019587
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr3_+_80556668 | 0.09 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr14_+_86652365 | 0.09 |
ENSRNOT00000077428
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr10_+_36715565 | 0.09 |
ENSRNOT00000005048
|
Clk4
|
CDC-like kinase 4 |
| chr1_+_256382791 | 0.09 |
ENSRNOT00000022549
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chrX_+_111735820 | 0.09 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr16_-_15798974 | 0.09 |
ENSRNOT00000046842
ENSRNOT00000065946 |
Nrg3
|
neuregulin 3 |
| chr7_+_72985495 | 0.09 |
ENSRNOT00000008361
|
Matn2
|
matrilin 2 |
| chr7_-_123621102 | 0.09 |
ENSRNOT00000046024
|
Cyp2d5
|
cytochrome P450, family 2, subfamily d, polypeptide 5 |
| chr13_-_103080920 | 0.09 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr1_-_222350173 | 0.09 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr2_-_200003443 | 0.09 |
ENSRNOT00000024900
ENSRNOT00000088041 |
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr17_+_22620721 | 0.09 |
ENSRNOT00000019478
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr1_-_167700332 | 0.09 |
ENSRNOT00000092890
|
Trim21
|
tripartite motif-containing 21 |
| chr3_-_48372583 | 0.09 |
ENSRNOT00000040482
ENSRNOT00000077788 ENSRNOT00000085426 |
Dpp4
|
dipeptidylpeptidase 4 |
| chr4_+_174692331 | 0.09 |
ENSRNOT00000011770
|
Plekha5
|
pleckstrin homology domain containing A5 |
| chr16_+_39909270 | 0.09 |
ENSRNOT00000081994
|
Wdr17
|
WD repeat domain 17 |
| chr19_+_10563423 | 0.09 |
ENSRNOT00000021037
|
Dok4
|
docking protein 4 |
| chr5_-_79222687 | 0.08 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr2_-_62634785 | 0.08 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr10_-_54467956 | 0.08 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chr11_+_61661310 | 0.08 |
ENSRNOT00000093325
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr3_-_39596718 | 0.08 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr8_+_115546712 | 0.08 |
ENSRNOT00000079801
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr8_-_78655856 | 0.08 |
ENSRNOT00000081185
|
Tcf12
|
transcription factor 12 |
| chr9_-_93404883 | 0.08 |
ENSRNOT00000025024
|
Nmur1
|
neuromedin U receptor 1 |
| chr1_+_214562897 | 0.08 |
ENSRNOT00000085125
|
Ap2a2
|
adaptor-related protein complex 2, alpha 2 subunit |
| chr9_+_12114977 | 0.08 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr5_-_57632177 | 0.08 |
ENSRNOT00000080787
ENSRNOT00000092581 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr11_+_61531416 | 0.08 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr10_+_57218087 | 0.08 |
ENSRNOT00000089853
|
Mink1
|
misshapen-like kinase 1 |
| chr2_-_22798214 | 0.08 |
ENSRNOT00000016135
|
Papd4
|
poly(A) RNA polymerase D4, non-canonical |
| chr20_-_8202924 | 0.08 |
ENSRNOT00000071399
|
Tmem217
|
transmembrane protein 217 |
| chr11_-_62451149 | 0.08 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr10_+_15183803 | 0.07 |
ENSRNOT00000039777
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr1_+_66898946 | 0.07 |
ENSRNOT00000074063
|
Zfp551
|
zinc finger protein 551 |
| chr9_+_118586179 | 0.07 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chr2_-_39003552 | 0.07 |
ENSRNOT00000078884
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr10_-_70802782 | 0.07 |
ENSRNOT00000045867
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr2_+_45668969 | 0.07 |
ENSRNOT00000014761
ENSRNOT00000071353 |
Arl15
|
ADP-ribosylation factor like GTPase 15 |
| chr16_-_3139177 | 0.07 |
ENSRNOT00000039311
|
Ccdc66
|
coiled-coil domain containing 66 |
| chr13_-_88642011 | 0.07 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr4_-_119591817 | 0.07 |
ENSRNOT00000048768
|
AC097129.1
|
|
| chr4_-_161681660 | 0.07 |
ENSRNOT00000039086
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chrX_+_88298266 | 0.07 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr14_-_3359300 | 0.07 |
ENSRNOT00000080875
|
Lpcat2b
|
lysophosphatidylcholine acyltransferase 2b |
| chr5_-_147412705 | 0.07 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr8_+_71167305 | 0.07 |
ENSRNOT00000021337
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr12_-_2555164 | 0.07 |
ENSRNOT00000084460
ENSRNOT00000061821 |
Map2k7
|
mitogen activated protein kinase kinase 7 |
| chr9_+_2202511 | 0.07 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr17_-_18590536 | 0.07 |
ENSRNOT00000078992
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr16_-_70705128 | 0.07 |
ENSRNOT00000071258
|
Rnf170
|
ring finger protein 170 |
| chr19_+_21218465 | 0.07 |
ENSRNOT00000049285
|
N4bp1
|
Nedd4 binding protein 1 |
| chr1_-_266074181 | 0.07 |
ENSRNOT00000026378
|
Psd
|
pleckstrin and Sec7 domain containing |
| chr3_+_138398011 | 0.07 |
ENSRNOT00000038865
|
Mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr12_-_46889082 | 0.07 |
ENSRNOT00000001525
|
Pla2g1b
|
phospholipase A2 group IB |
| chr2_-_234296145 | 0.07 |
ENSRNOT00000014155
|
Elovl6
|
ELOVL fatty acid elongase 6 |
| chr7_-_140502441 | 0.07 |
ENSRNOT00000089544
|
Kmt2d
|
lysine methyltransferase 2D |
| chr8_-_130338325 | 0.07 |
ENSRNOT00000071697
|
Sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr1_-_13915594 | 0.07 |
ENSRNOT00000015927
|
Arfgef3
|
ARFGEF family member 3 |
| chr12_-_37398329 | 0.07 |
ENSRNOT00000083401
|
Tctn2
|
tectonic family member 2 |
| chr1_-_191651628 | 0.07 |
ENSRNOT00000055048
|
Usp31
|
ubiquitin specific peptidase 31 |
| chr7_+_127964752 | 0.06 |
ENSRNOT00000038168
|
RGD1560617
|
hypothetical gene supported by NM_053561; AF062594 |
| chr18_-_62614725 | 0.06 |
ENSRNOT00000025223
|
Mc4r
|
melanocortin 4 receptor |
| chr12_+_45727112 | 0.06 |
ENSRNOT00000001507
|
Srrm4
|
serine/arginine repetitive matrix 4 |
| chr3_+_161272385 | 0.06 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chr9_+_82718709 | 0.06 |
ENSRNOT00000027256
ENSRNOT00000080524 |
Stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr15_+_37171052 | 0.06 |
ENSRNOT00000011684
|
Zmym2
|
zinc finger MYM-type containing 2 |
| chr7_-_70577147 | 0.06 |
ENSRNOT00000008854
|
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr1_-_90520012 | 0.06 |
ENSRNOT00000028698
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr12_-_22245100 | 0.06 |
ENSRNOT00000001912
|
Gigyf1
|
GRB10 interacting GYF protein 1 |
| chr14_+_1355004 | 0.06 |
ENSRNOT00000043131
|
AABR07014114.1
|
|
| chr4_+_170518673 | 0.06 |
ENSRNOT00000011803
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr10_+_39875371 | 0.06 |
ENSRNOT00000013481
|
Rapgef6
|
Rap guanine nucleotide exchange factor 6 |
| chr1_+_104635989 | 0.06 |
ENSRNOT00000078477
|
Nav2
|
neuron navigator 2 |
| chr11_-_70833577 | 0.06 |
ENSRNOT00000002428
|
Osbpl11
|
oxysterol binding protein-like 11 |
| chr17_-_80320681 | 0.06 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr1_+_85162452 | 0.06 |
ENSRNOT00000093347
|
Pak4
|
p21 (RAC1) activated kinase 4 |
| chr7_+_110031696 | 0.06 |
ENSRNOT00000012753
|
Khdrbs3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr13_-_48286720 | 0.06 |
ENSRNOT00000008976
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr13_-_39643361 | 0.06 |
ENSRNOT00000003527
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr7_+_11490852 | 0.06 |
ENSRNOT00000044484
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3 |
| chr5_+_150754021 | 0.06 |
ENSRNOT00000017687
|
Ptafr
|
platelet-activating factor receptor |
| chr18_+_60496778 | 0.06 |
ENSRNOT00000088624
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr9_+_81783349 | 0.06 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr20_-_12820466 | 0.06 |
ENSRNOT00000001699
|
Ftcd
|
formimidoyltransferase cyclodeaminase |
| chr3_+_103773459 | 0.06 |
ENSRNOT00000079727
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr9_+_18564927 | 0.06 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr11_+_61321459 | 0.06 |
ENSRNOT00000002759
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr4_+_158224000 | 0.06 |
ENSRNOT00000084240
ENSRNOT00000078495 |
Ano2
|
anoctamin 2 |
| chr13_-_105141030 | 0.06 |
ENSRNOT00000003313
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr2_+_187893875 | 0.05 |
ENSRNOT00000093014
|
Mex3a
|
mex-3 RNA binding family member A |
| chr3_+_59819157 | 0.05 |
ENSRNOT00000040114
|
LOC103695172
|
uncharacterized LOC103695172 |
| chr1_-_277355075 | 0.05 |
ENSRNOT00000036321
|
Dclre1a
|
DNA cross-link repair 1A |
| chr18_+_30904498 | 0.05 |
ENSRNOT00000026969
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr3_-_13525983 | 0.05 |
ENSRNOT00000082036
|
Pbx3
|
PBX homeobox 3 |
| chr3_+_63379031 | 0.05 |
ENSRNOT00000068199
|
Osbpl6
|
oxysterol binding protein-like 6 |
| chr8_+_50525091 | 0.05 |
ENSRNOT00000074357
|
Apoa1
|
apolipoprotein A1 |
| chr7_-_84023316 | 0.05 |
ENSRNOT00000005556
|
Kcnv1
|
potassium voltage-gated channel modifier subfamily V member 1 |
| chr19_-_56772904 | 0.05 |
ENSRNOT00000024232
|
Abcb10
|
ATP binding cassette subfamily B member 10 |
| chr17_-_44640092 | 0.05 |
ENSRNOT00000077628
|
Zfp184
|
zinc finger protein 184 |
| chr4_+_96562725 | 0.05 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr1_-_37726151 | 0.05 |
ENSRNOT00000071842
|
RGD1308544
|
LOC361192 |
| chr19_-_22281778 | 0.05 |
ENSRNOT00000049624
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_-_88112683 | 0.05 |
ENSRNOT00000090615
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr18_+_59748444 | 0.05 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr7_-_120770435 | 0.05 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chrX_+_124321551 | 0.05 |
ENSRNOT00000074486
|
LOC100910807
|
transcriptional regulator Kaiso-like |
| chr4_+_113101100 | 0.05 |
ENSRNOT00000084897
|
AABR07061261.1
|
|
| chr7_+_27620458 | 0.05 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr4_-_150471806 | 0.05 |
ENSRNOT00000008741
ENSRNOT00000076557 |
Bms1
|
BMS1 ribosome biogenesis factor |
| chr14_-_114649173 | 0.05 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr7_+_15422479 | 0.05 |
ENSRNOT00000066520
|
Zfp563
|
zinc finger protein 563 |
| chr1_+_197839430 | 0.05 |
ENSRNOT00000025043
|
Rabep2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr17_-_52569036 | 0.05 |
ENSRNOT00000019396
|
Gli3
|
GLI family zinc finger 3 |
| chr9_+_10471742 | 0.05 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chrX_+_35869538 | 0.05 |
ENSRNOT00000058947
|
Ppef1
|
protein phosphatase with EF-hand domain 1 |
| chr7_-_104749552 | 0.05 |
ENSRNOT00000079981
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr3_+_54253949 | 0.05 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr1_+_219144205 | 0.05 |
ENSRNOT00000083942
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr1_-_85317968 | 0.05 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr18_-_56325095 | 0.05 |
ENSRNOT00000025209
|
Slc6a7
|
solute carrier family 6 member 7 |
| chr11_+_57430166 | 0.05 |
ENSRNOT00000093201
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr6_+_73358112 | 0.05 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr12_-_36398206 | 0.05 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr8_+_58120179 | 0.05 |
ENSRNOT00000049076
ENSRNOT00000090114 |
Npat
|
nuclear protein, co-activator of histone transcription |
| chr5_-_57607725 | 0.05 |
ENSRNOT00000080295
|
Ubap2
|
ubiquitin-associated protein 2 |
| chr2_-_22744407 | 0.05 |
ENSRNOT00000073710
|
Cmya5
|
cardiomyopathy associated 5 |
| chr5_+_172881813 | 0.05 |
ENSRNOT00000067227
|
AC130035.1
|
|
| chr8_+_71168097 | 0.05 |
ENSRNOT00000081375
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr13_+_110920830 | 0.05 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr2_-_195678848 | 0.05 |
ENSRNOT00000028303
ENSRNOT00000075569 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr11_-_87158597 | 0.05 |
ENSRNOT00000002517
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr1_+_256226179 | 0.05 |
ENSRNOT00000054748
|
Exoc6
|
exocyst complex component 6 |
| chr9_-_70787913 | 0.05 |
ENSRNOT00000072007
ENSRNOT00000017901 |
Klf7
Klf7
|
Kruppel like factor 7 Kruppel like factor 7 |
| chr1_+_7252349 | 0.05 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chrX_-_142248369 | 0.05 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr4_-_150829741 | 0.05 |
ENSRNOT00000051846
ENSRNOT00000052017 |
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr6_-_61405195 | 0.04 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr1_+_85192174 | 0.04 |
ENSRNOT00000026966
|
Pak4
|
p21 (RAC1) activated kinase 4 |
| chr4_+_123801174 | 0.04 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr2_-_199354793 | 0.04 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr3_+_33440191 | 0.04 |
ENSRNOT00000034632
ENSRNOT00000092907 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr14_+_42015347 | 0.04 |
ENSRNOT00000044017
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr5_+_123905166 | 0.04 |
ENSRNOT00000082021
|
Dab1
|
DAB1, reelin adaptor protein |
| chr4_+_157659147 | 0.04 |
ENSRNOT00000048379
|
Iffo1
|
intermediate filament family orphan 1 |
| chr4_-_119131202 | 0.04 |
ENSRNOT00000011675
|
Antxr1
|
anthrax toxin receptor 1 |
| chr12_-_24537313 | 0.04 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr8_-_47339343 | 0.04 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr20_-_3374344 | 0.04 |
ENSRNOT00000082999
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr5_+_148200370 | 0.04 |
ENSRNOT00000064948
|
Adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr3_+_71114100 | 0.04 |
ENSRNOT00000088549
ENSRNOT00000006961 |
Itgav
|
integrin subunit alpha V |
| chr9_-_10471009 | 0.04 |
ENSRNOT00000072868
|
Safb
|
scaffold attachment factor B |
| chr3_+_80676820 | 0.04 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr1_+_227757425 | 0.04 |
ENSRNOT00000032937
ENSRNOT00000049574 |
Ms4a6bl
|
membrane-spanning 4-domains, subfamily A, member 6B-like |
| chr1_+_48433079 | 0.04 |
ENSRNOT00000037369
|
Slc22a3
|
solute carrier family 22 member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1i2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.1 | GO:1905006 | positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.0 | 0.1 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.0 | 0.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:0043132 | phospholipid transfer to membrane(GO:0006649) NAD transport(GO:0043132) regulation of bleb assembly(GO:1904170) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0061744 | motor behavior(GO:0061744) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.0 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.0 | 0.0 | GO:1901726 | negative regulation of histone deacetylase activity(GO:1901726) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.0 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:0021812 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.0 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.0 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0042447 | phenol-containing compound catabolic process(GO:0019336) hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.1 | GO:0006863 | purine nucleobase transport(GO:0006863) nucleobase transport(GO:0015851) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0033222 | xylose binding(GO:0033222) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |