Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
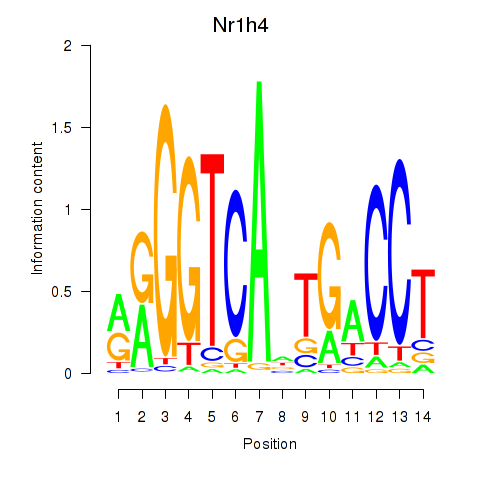
Results for Nr1h4
Z-value: 0.36
Transcription factors associated with Nr1h4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1h4
|
ENSRNOG00000007197 | nuclear receptor subfamily 1, group H, member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1h4 | rn6_v1_chr7_-_30105132_30105132 | 0.91 | 3.1e-02 | Click! |
Activity profile of Nr1h4 motif
Sorted Z-values of Nr1h4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_55178289 | 0.20 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr15_-_25521393 | 0.19 |
ENSRNOT00000085910
|
Otx2
|
orthodenticle homeobox 2 |
| chr14_+_83724933 | 0.14 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr1_-_89488223 | 0.13 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr2_+_187740531 | 0.12 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr15_-_25505691 | 0.12 |
ENSRNOT00000074488
|
LOC100911492
|
homeobox protein OTX2-like |
| chr7_+_130542202 | 0.12 |
ENSRNOT00000079501
ENSRNOT00000045647 |
Acr
|
acrosin |
| chr12_+_47024442 | 0.11 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr8_+_23113048 | 0.11 |
ENSRNOT00000029577
|
Cnn1
|
calponin 1 |
| chr19_-_15733412 | 0.10 |
ENSRNOT00000014831
|
Irx6
|
iroquois homeobox 6 |
| chr4_+_100407658 | 0.10 |
ENSRNOT00000018562
|
Capg
|
capping actin protein, gelsolin like |
| chr2_-_187401786 | 0.10 |
ENSRNOT00000025624
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr7_-_12609694 | 0.10 |
ENSRNOT00000093287
ENSRNOT00000079485 |
Kiss1r
|
KISS1 receptor |
| chr8_-_23014499 | 0.09 |
ENSRNOT00000017820
|
Ccdc151
|
coiled-coil domain containing 151 |
| chr10_+_13836128 | 0.09 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr7_-_12609868 | 0.09 |
ENSRNOT00000016396
|
Kiss1r
|
KISS1 receptor |
| chr18_+_57011575 | 0.08 |
ENSRNOT00000026679
|
Il17b
|
interleukin 17B |
| chr3_+_118317761 | 0.08 |
ENSRNOT00000012700
|
Fgf7
|
fibroblast growth factor 7 |
| chr9_+_69953440 | 0.07 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr15_+_4475051 | 0.07 |
ENSRNOT00000008461
|
Kcnk16
|
potassium two pore domain channel subfamily K member 16 |
| chr4_+_61850348 | 0.07 |
ENSRNOT00000013423
|
Akr1b7
|
aldo-keto reductase family 1, member B7 |
| chr14_-_83776863 | 0.07 |
ENSRNOT00000026481
|
Smtn
|
smoothelin |
| chr9_+_27068443 | 0.07 |
ENSRNOT00000017393
|
Efhc1
|
EF-hand domain containing 1 |
| chr20_+_5049496 | 0.07 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr10_+_14598014 | 0.07 |
ENSRNOT00000068336
|
Tsr3
|
TSR3, 20S rRNA accumulation |
| chr10_-_97756521 | 0.06 |
ENSRNOT00000045902
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr13_+_71086745 | 0.06 |
ENSRNOT00000083366
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr1_+_221773254 | 0.06 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr10_-_19715832 | 0.06 |
ENSRNOT00000040278
|
RGD1564698
|
similar to ribosomal protein S10 |
| chr14_+_2325308 | 0.06 |
ENSRNOT00000000072
|
Atp5i
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr17_-_9234766 | 0.06 |
ENSRNOT00000015893
|
AABR07026997.1
|
|
| chr9_+_74124016 | 0.06 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr15_+_44441856 | 0.06 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr12_-_30566032 | 0.06 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr1_-_64099277 | 0.05 |
ENSRNOT00000084846
|
Tsen34
|
tRNA splicing endonuclease subunit 34 |
| chr8_+_116857684 | 0.05 |
ENSRNOT00000026711
|
Mst1
|
macrophage stimulating 1 |
| chr6_-_136550371 | 0.05 |
ENSRNOT00000065971
|
Rd3l
|
retinal degeneration 3-like |
| chr1_+_225037737 | 0.05 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr20_+_5441876 | 0.05 |
ENSRNOT00000092476
|
Rps18
|
ribosomal protein S18 |
| chr10_+_59539405 | 0.05 |
ENSRNOT00000077107
|
Itgae
|
integrin subunit alpha E |
| chr1_-_170431073 | 0.05 |
ENSRNOT00000024710
|
Hpx
|
hemopexin |
| chr10_-_56223245 | 0.05 |
ENSRNOT00000015248
|
Shbg
|
sex hormone binding globulin |
| chr9_-_64096265 | 0.05 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr9_+_64095978 | 0.05 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr14_+_88549947 | 0.04 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr13_-_89619398 | 0.04 |
ENSRNOT00000058423
|
Ndufs2
|
NADH dehydrogenase (ubiquinone) Fe-S protein 2 |
| chr5_+_136683592 | 0.04 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr16_+_6903212 | 0.04 |
ENSRNOT00000023314
|
Tmem110
|
transmembrane protein 110 |
| chr12_+_2180150 | 0.04 |
ENSRNOT00000001322
|
Stxbp2
|
syntaxin binding protein 2 |
| chr3_-_47025128 | 0.04 |
ENSRNOT00000011682
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr19_+_24786628 | 0.04 |
ENSRNOT00000005374
|
Gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr3_-_55587946 | 0.04 |
ENSRNOT00000075107
|
Abcb11
|
ATP binding cassette subfamily B member 11 |
| chr9_+_61655963 | 0.04 |
ENSRNOT00000040461
|
Coq10b
|
coenzyme Q10B |
| chr7_+_70580198 | 0.04 |
ENSRNOT00000083472
ENSRNOT00000008941 |
Ddit3
|
DNA-damage inducible transcript 3 |
| chr7_-_12882753 | 0.04 |
ENSRNOT00000011275
ENSRNOT00000044275 |
Bsg
|
basigin |
| chr6_+_107245820 | 0.04 |
ENSRNOT00000012757
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr3_-_11810901 | 0.04 |
ENSRNOT00000071233
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr7_-_143837780 | 0.03 |
ENSRNOT00000016642
|
Itgb7
|
integrin subunit beta 7 |
| chr18_-_62965538 | 0.03 |
ENSRNOT00000025164
|
Mppe1
|
metallophosphoesterase 1 |
| chr14_+_24129592 | 0.03 |
ENSRNOT00000040647
|
LOC689899
|
similar to 60S ribosomal protein L23a |
| chr6_+_124123228 | 0.03 |
ENSRNOT00000005329
|
Psmc1
|
proteasome 26S subunit, ATPase 1 |
| chr2_+_248649441 | 0.03 |
ENSRNOT00000067165
|
Kyat3
|
kynurenine aminotransferase 3 |
| chr16_-_7007287 | 0.03 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr18_+_16616937 | 0.03 |
ENSRNOT00000093641
|
Mocos
|
molybdenum cofactor sulfurase |
| chrX_+_80359555 | 0.03 |
ENSRNOT00000030692
|
LOC681325
|
hypothetical protein LOC681325 |
| chr9_+_43830630 | 0.03 |
ENSRNOT00000084742
|
Cnga3
|
cyclic nucleotide gated channel alpha 3 |
| chr6_-_10899200 | 0.03 |
ENSRNOT00000089104
|
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr8_-_25829569 | 0.03 |
ENSRNOT00000071884
|
Dpy19l2
|
dpy-19 like 2 |
| chr12_+_22727335 | 0.03 |
ENSRNOT00000077293
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr7_-_127081704 | 0.03 |
ENSRNOT00000055893
|
LOC108351584
|
uncharacterized LOC108351584 |
| chr6_+_107550904 | 0.03 |
ENSRNOT00000013760
|
Acot5
|
acyl-CoA thioesterase 5 |
| chr4_+_100277391 | 0.03 |
ENSRNOT00000086820
ENSRNOT00000084281 ENSRNOT00000017927 |
Ggcx
|
gamma-glutamyl carboxylase |
| chr2_-_198439454 | 0.03 |
ENSRNOT00000028780
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr20_+_5442390 | 0.03 |
ENSRNOT00000037499
|
Rps18
|
ribosomal protein S18 |
| chr9_+_82647071 | 0.02 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr3_-_153468221 | 0.02 |
ENSRNOT00000010443
|
Ghrh
|
growth hormone releasing hormone |
| chr1_+_198409360 | 0.02 |
ENSRNOT00000013691
ENSRNOT00000091295 |
Cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr3_+_102925471 | 0.02 |
ENSRNOT00000040364
|
Olr772
|
olfactory receptor 772 |
| chr7_+_117326279 | 0.02 |
ENSRNOT00000050522
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr18_-_36579403 | 0.02 |
ENSRNOT00000025284
|
Lars
|
leucyl-tRNA synthetase |
| chr1_-_80617057 | 0.02 |
ENSRNOT00000080453
|
Apoe
|
apolipoprotein E |
| chr10_+_56576428 | 0.02 |
ENSRNOT00000079237
ENSRNOT00000023291 |
Cldn7
|
claudin 7 |
| chr8_+_117455262 | 0.02 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr1_-_170628915 | 0.02 |
ENSRNOT00000042865
|
Dchs1
|
dachsous cadherin-related 1 |
| chr16_+_35934970 | 0.02 |
ENSRNOT00000084707
ENSRNOT00000016474 |
Galnt7
|
polypeptide N-acetylgalactosaminyltransferase 7 |
| chr17_-_42127678 | 0.02 |
ENSRNOT00000024196
|
Gpld1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr17_+_81280898 | 0.02 |
ENSRNOT00000057785
|
Tmem236
|
transmembrane protein 236 |
| chr15_-_51839341 | 0.02 |
ENSRNOT00000011277
|
RGD1308117
|
similar to 9930012K11Rik protein |
| chr3_-_151371501 | 0.02 |
ENSRNOT00000073178
|
Fam83c
|
family with sequence similarity 83, member C |
| chr15_-_52712995 | 0.02 |
ENSRNOT00000039731
|
LOC100359986
|
ribosomal protein L31-like |
| chr1_+_72692105 | 0.02 |
ENSRNOT00000032439
|
Tmem150b
|
transmembrane protein 150B |
| chr1_-_219259448 | 0.02 |
ENSRNOT00000024517
|
Ndufv1
|
NADH:ubiquinone oxidoreductase core subunit V1 |
| chr14_-_33580566 | 0.02 |
ENSRNOT00000063942
|
Paics
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr12_+_9034308 | 0.02 |
ENSRNOT00000001248
|
Flt1
|
FMS-related tyrosine kinase 1 |
| chr5_+_36024649 | 0.02 |
ENSRNOT00000013384
|
Coq3
|
coenzyme Q3 methyltransferase |
| chr8_+_23014956 | 0.02 |
ENSRNOT00000018009
|
Prkcsh
|
protein kinase C substrate 80K-H |
| chr13_-_74740458 | 0.02 |
ENSRNOT00000006548
|
Tex35
|
testis expressed 35 |
| chr6_+_126766967 | 0.02 |
ENSRNOT00000052296
|
Cox8c
|
cytochrome c oxidase subunit 8C |
| chr10_+_37582706 | 0.01 |
ENSRNOT00000007630
|
RGD1306484
|
similar to growth and transformation-dependent protein |
| chr1_+_100470722 | 0.01 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr3_-_81911197 | 0.01 |
ENSRNOT00000066526
|
Prdm11
|
PR/SET domain 11 |
| chrX_+_21696772 | 0.01 |
ENSRNOT00000043559
|
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr5_+_151776004 | 0.01 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr10_-_46582854 | 0.01 |
ENSRNOT00000004753
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr1_+_100447998 | 0.01 |
ENSRNOT00000026259
|
Lrrc4b
|
leucine rich repeat containing 4B |
| chr3_+_4233111 | 0.01 |
ENSRNOT00000042910
|
Lcn1
|
lipocalin 1 |
| chr9_+_82700468 | 0.01 |
ENSRNOT00000027227
|
Inha
|
inhibin alpha subunit |
| chr1_+_219833299 | 0.01 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chrX_-_15768522 | 0.01 |
ENSRNOT00000077391
|
Foxp3
|
forkhead box P3 |
| chr5_-_154319629 | 0.01 |
ENSRNOT00000014415
|
Lypla2
|
lysophospholipase II |
| chr1_-_98486976 | 0.01 |
ENSRNOT00000024083
|
Etfb
|
electron transfer flavoprotein beta subunit |
| chr10_-_109913879 | 0.01 |
ENSRNOT00000072947
|
LOC688320
|
similar to Lung carbonyl reductase [NADPH] (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27) |
| chr1_+_89215266 | 0.01 |
ENSRNOT00000093612
ENSRNOT00000084799 |
Dmkn
|
dermokine |
| chr14_+_36216002 | 0.01 |
ENSRNOT00000038506
|
Scfd2
|
sec1 family domain containing 2 |
| chr6_-_59531574 | 0.01 |
ENSRNOT00000042029
|
RGD1560073
|
similar to ribosomal protein S10 |
| chr8_+_115151627 | 0.01 |
ENSRNOT00000064252
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr7_-_102298522 | 0.00 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chr3_+_7051695 | 0.00 |
ENSRNOT00000013548
|
Mrps2
|
mitochondrial ribosomal protein S2 |
| chr7_-_123655896 | 0.00 |
ENSRNOT00000012413
|
Cyp2d2
|
cytochrome P450, family 2, subfamily d, polypeptide 2 |
| chr14_+_79261092 | 0.00 |
ENSRNOT00000029191
|
LOC680039
|
hypothetical protein LOC680039 |
| chr16_-_7007051 | 0.00 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr4_-_157381105 | 0.00 |
ENSRNOT00000021670
|
Gpr162
|
G protein-coupled receptor 162 |
| chr2_+_20857202 | 0.00 |
ENSRNOT00000078919
|
Acot12
|
acyl-CoA thioesterase 12 |
| chr12_-_12907771 | 0.00 |
ENSRNOT00000001412
|
Cyth3
|
cytohesin 3 |
| chr4_-_117790350 | 0.00 |
ENSRNOT00000088557
|
Nat8b
|
N-acetyltransferase 8B |
| chr1_+_264741911 | 0.00 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr12_+_37490584 | 0.00 |
ENSRNOT00000001401
ENSRNOT00000090831 |
Rilpl1
|
Rab interacting lysosomal protein-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1h4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042706 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.0 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.0 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.0 | GO:1990622 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |