Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
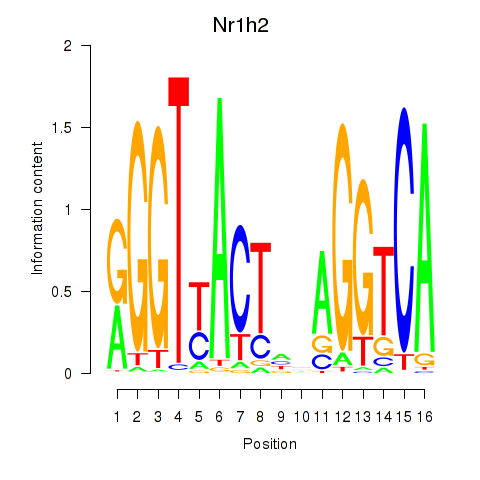
Results for Nr1h2
Z-value: 0.36
Transcription factors associated with Nr1h2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr1h2
|
ENSRNOG00000019812 | nuclear receptor subfamily 1, group H, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr1h2 | rn6_v1_chr1_-_100559942_100559942 | 0.76 | 1.3e-01 | Click! |
Activity profile of Nr1h2 motif
Sorted Z-values of Nr1h2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_10047507 | 0.19 |
ENSRNOT00000072118
|
Alkbh7
|
alkB homolog 7 |
| chr12_-_38995570 | 0.18 |
ENSRNOT00000001806
|
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr1_+_88182585 | 0.15 |
ENSRNOT00000044145
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr1_+_87180624 | 0.12 |
ENSRNOT00000056933
ENSRNOT00000027987 |
LOC103689986
|
protein YIF1B |
| chr1_+_88182155 | 0.12 |
ENSRNOT00000083176
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr1_-_80744831 | 0.12 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr1_+_81373340 | 0.12 |
ENSRNOT00000026814
|
Zfp428
|
zinc finger protein 428 |
| chr10_-_46582854 | 0.11 |
ENSRNOT00000004753
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr12_+_18516946 | 0.10 |
ENSRNOT00000029485
|
Dhrsx
|
dehydrogenase/reductase X-linked |
| chrX_-_13116743 | 0.09 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr16_+_19792838 | 0.09 |
ENSRNOT00000022938
|
Babam1
|
BRISC and BRCA1 A complex member 1 |
| chr20_+_7136007 | 0.08 |
ENSRNOT00000000580
|
Hmga1
|
high mobility group AT-hook 1 |
| chr4_+_83391283 | 0.08 |
ENSRNOT00000031365
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr5_-_153790157 | 0.07 |
ENSRNOT00000025051
|
Rcan3
|
RCAN family member 3 |
| chr6_-_7058314 | 0.07 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr10_-_89084885 | 0.07 |
ENSRNOT00000027452
|
Plekhh3
|
pleckstrin homology, MyTH4 and FERM domain containing H3 |
| chr4_-_165629996 | 0.07 |
ENSRNOT00000068185
ENSRNOT00000007427 |
Ybx3
|
Y box binding protein 3 |
| chr13_+_52553775 | 0.06 |
ENSRNOT00000011991
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr10_+_86860685 | 0.06 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chrX_+_13117239 | 0.06 |
ENSRNOT00000088998
|
AABR07037112.1
|
|
| chr7_-_11963268 | 0.06 |
ENSRNOT00000080342
ENSRNOT00000025266 |
Csnk1g2
|
casein kinase 1, gamma 2 |
| chr9_+_45672157 | 0.06 |
ENSRNOT00000017882
|
Pdcl3
|
phosducin-like 3 |
| chr12_-_46889082 | 0.06 |
ENSRNOT00000001525
|
Pla2g1b
|
phospholipase A2 group IB |
| chr3_+_172195844 | 0.05 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr5_-_151768123 | 0.05 |
ENSRNOT00000079380
|
Nudc
|
nuclear distribution C, dynein complex regulator |
| chr20_+_44680449 | 0.05 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr1_-_221507181 | 0.05 |
ENSRNOT00000093048
|
Arl2
|
ADP-ribosylation factor like GTPase 2 |
| chr12_+_52356832 | 0.05 |
ENSRNOT00000050252
|
Fbrsl1
|
fibrosin-like 1 |
| chr2_-_140464607 | 0.05 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr10_-_56558487 | 0.05 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr2_+_30685840 | 0.05 |
ENSRNOT00000031385
|
Ccdc125
|
coiled-coil domain containing 125 |
| chr20_-_22459025 | 0.05 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr10_-_62273119 | 0.04 |
ENSRNOT00000004322
|
Serpinf2
|
serpin family F member 2 |
| chr1_-_80689171 | 0.04 |
ENSRNOT00000045574
|
Bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr4_+_84597323 | 0.04 |
ENSRNOT00000074054
ENSRNOT00000012755 |
Wipf3
|
WAS/WASL interacting protein family, member 3 |
| chr5_+_157423213 | 0.04 |
ENSRNOT00000023431
|
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr10_+_40438356 | 0.04 |
ENSRNOT00000078910
|
Gm2a
|
GM2 ganglioside activator |
| chr7_+_141034982 | 0.04 |
ENSRNOT00000091050
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr17_-_43675934 | 0.04 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr18_+_3565166 | 0.04 |
ENSRNOT00000039377
|
Riok3
|
RIO kinase 3 |
| chr12_-_22006533 | 0.03 |
ENSRNOT00000059593
|
LOC100362783
|
Uncharacterized protein C7orf61 homolog |
| chr9_-_71313940 | 0.03 |
ENSRNOT00000019670
|
Mettl21a
|
methyltransferase like 21A |
| chr13_-_92089980 | 0.03 |
ENSRNOT00000036040
|
Mnda
|
myeloid cell nuclear differentiation antigen |
| chr3_+_150910398 | 0.03 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr7_-_144269486 | 0.03 |
ENSRNOT00000090051
|
Atp5g2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr3_-_94808861 | 0.03 |
ENSRNOT00000038464
|
Prrg4
|
proline rich and Gla domain 4 |
| chr2_-_196415530 | 0.03 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr20_-_7208292 | 0.03 |
ENSRNOT00000083169
|
Nudt3
|
nudix hydrolase 3 |
| chr5_-_138470096 | 0.03 |
ENSRNOT00000011392
|
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr1_-_188407132 | 0.03 |
ENSRNOT00000073233
|
Gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr15_-_56970365 | 0.03 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr7_-_58286770 | 0.03 |
ENSRNOT00000005258
|
Rab21
|
RAB21, member RAS oncogene family |
| chrX_+_22788660 | 0.03 |
ENSRNOT00000071886
|
AABR07037510.1
|
|
| chr14_+_80195715 | 0.03 |
ENSRNOT00000010784
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr1_-_24191908 | 0.02 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr19_+_25969255 | 0.02 |
ENSRNOT00000004370
|
Farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr1_-_199177365 | 0.02 |
ENSRNOT00000041840
|
Ctf2
|
cardiotrophin 2 |
| chr3_-_123206828 | 0.02 |
ENSRNOT00000030192
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr1_+_140477868 | 0.02 |
ENSRNOT00000025008
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr1_-_153096269 | 0.02 |
ENSRNOT00000022578
|
Tmem135
|
transmembrane protein 135 |
| chr5_-_157758084 | 0.02 |
ENSRNOT00000083341
|
Pqlc2
|
PQ loop repeat containing 2 |
| chr3_-_5452156 | 0.02 |
ENSRNOT00000006699
|
Surf6
|
surfeit 6 |
| chr4_-_170167633 | 0.02 |
ENSRNOT00000007402
|
Wbp11l1
|
WW domain-binding protein 11-like 1 |
| chr12_+_41543696 | 0.02 |
ENSRNOT00000079639
|
Tpcn1
|
two pore segment channel 1 |
| chr11_-_87236445 | 0.02 |
ENSRNOT00000057806
|
Tssk2
|
testis-specific serine kinase 2 |
| chr1_-_140477796 | 0.02 |
ENSRNOT00000067172
|
Mrpl46
|
mitochondrial ribosomal protein L46 |
| chr6_+_145770662 | 0.02 |
ENSRNOT00000088969
|
Cdca7l
|
cell division cycle associated 7 like |
| chr11_-_55240241 | 0.02 |
ENSRNOT00000064614
|
Dppa4
|
developmental pluripotency associated 4 |
| chr2_+_187740531 | 0.02 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr9_+_81631409 | 0.02 |
ENSRNOT00000089580
|
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr13_+_34483876 | 0.02 |
ENSRNOT00000092998
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr4_-_145390447 | 0.02 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr20_+_28920616 | 0.02 |
ENSRNOT00000070897
|
P4ha1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr1_+_227670159 | 0.02 |
ENSRNOT00000072077
|
Ms4a6c
|
membrane-spanning 4-domains, subfamily A, member 6C |
| chr16_+_20824864 | 0.02 |
ENSRNOT00000092293
ENSRNOT00000092446 |
Upf1
|
UPF1, RNA helicase and ATPase |
| chr1_+_247688789 | 0.02 |
ENSRNOT00000021783
|
Ric1
|
RIC1 homolog, RAB6A GEF complex partner 1 |
| chr1_-_216920625 | 0.02 |
ENSRNOT00000028119
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr3_+_38742072 | 0.02 |
ENSRNOT00000006774
|
Arl6ip6
|
ADP ribosylation factor like GTPase 6 interacting protein 6 |
| chr1_-_216744066 | 0.02 |
ENSRNOT00000087961
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr7_-_144837395 | 0.01 |
ENSRNOT00000089024
|
Cbx5
|
chromobox 5 |
| chr1_-_84254645 | 0.01 |
ENSRNOT00000077651
|
Sptbn4
|
spectrin, beta, non-erythrocytic 4 |
| chr11_-_61530830 | 0.01 |
ENSRNOT00000059666
ENSRNOT00000068345 |
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr13_-_90977734 | 0.01 |
ENSRNOT00000011869
|
Slamf8
|
SLAM family member 8 |
| chr1_-_72335855 | 0.01 |
ENSRNOT00000021613
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr20_+_21316826 | 0.01 |
ENSRNOT00000000785
|
RGD1306739
|
similar to RIKEN cDNA 1700040L02 |
| chr9_-_16647458 | 0.01 |
ENSRNOT00000024380
|
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr17_+_507377 | 0.01 |
ENSRNOT00000023638
|
Npepo
|
aminopeptidase O |
| chr9_+_53626592 | 0.01 |
ENSRNOT00000031267
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr19_-_10653800 | 0.01 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chr3_-_161322289 | 0.01 |
ENSRNOT00000022594
|
Pltp
|
phospholipid transfer protein |
| chr3_+_128756799 | 0.01 |
ENSRNOT00000049855
ENSRNOT00000042853 |
Plcb4
|
phospholipase C, beta 4 |
| chr8_+_61901128 | 0.01 |
ENSRNOT00000033209
|
LOC691110
|
similar to taste receptor protein 1 |
| chr14_+_11095163 | 0.01 |
ENSRNOT00000003076
|
Tmem150c
|
transmembrane protein 150C |
| chr7_-_11330278 | 0.01 |
ENSRNOT00000027730
|
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr1_+_244630377 | 0.01 |
ENSRNOT00000016740
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_6703979 | 0.01 |
ENSRNOT00000061246
|
Tex26
|
testis expressed 26 |
| chr9_+_16647598 | 0.01 |
ENSRNOT00000087413
|
Klc4
|
kinesin light chain 4 |
| chr4_+_162392869 | 0.01 |
ENSRNOT00000047790
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr10_-_15346236 | 0.01 |
ENSRNOT00000088104
ENSRNOT00000027430 |
Capn15
|
calpain 15 |
| chrX_+_26294066 | 0.01 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr3_-_9936352 | 0.01 |
ENSRNOT00000011224
ENSRNOT00000042798 |
Fnbp1
|
formin binding protein 1 |
| chr5_+_136406130 | 0.01 |
ENSRNOT00000093099
|
Eri3
|
ERI1 exoribonuclease family member 3 |
| chr12_+_10254951 | 0.01 |
ENSRNOT00000061129
|
Gpr12
|
G protein-coupled receptor 12 |
| chr6_-_129509720 | 0.01 |
ENSRNOT00000006131
|
Atg2b
|
autophagy related 2B |
| chr10_+_65507364 | 0.01 |
ENSRNOT00000016297
|
Sdf2
|
stromal cell derived factor 2 |
| chr4_-_117607428 | 0.01 |
ENSRNOT00000021243
|
LOC103690139
|
probable N-acetyltransferase CML6 |
| chr10_-_4910305 | 0.01 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr7_+_106535434 | 0.01 |
ENSRNOT00000045357
|
Efr3a
|
EFR3 homolog A |
| chr4_-_90882285 | 0.01 |
ENSRNOT00000039247
|
Snca
|
synuclein alpha |
| chrX_+_106823491 | 0.01 |
ENSRNOT00000045997
|
Bex3
|
brain expressed X-linked 3 |
| chr2_-_188561267 | 0.01 |
ENSRNOT00000089781
ENSRNOT00000092093 |
Trim46
|
tripartite motif-containing 46 |
| chr11_-_43992598 | 0.00 |
ENSRNOT00000002260
|
Cldnd1
|
claudin domain containing 1 |
| chr18_-_57866741 | 0.00 |
ENSRNOT00000025790
|
Fbxo38
|
F-box protein 38 |
| chr16_-_81797815 | 0.00 |
ENSRNOT00000026666
|
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr1_+_252375933 | 0.00 |
ENSRNOT00000078405
|
Lipn
|
lipase, family member N |
| chr10_-_82785142 | 0.00 |
ENSRNOT00000005381
|
Sgca
|
sarcoglycan, alpha |
| chr16_+_47368768 | 0.00 |
ENSRNOT00000017966
|
Wwc2
|
WW and C2 domain containing 2 |
| chr1_-_156296161 | 0.00 |
ENSRNOT00000025653
|
Crebzf
|
CREB/ATF bZIP transcription factor |
| chr13_-_111765944 | 0.00 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr4_-_117790350 | 0.00 |
ENSRNOT00000088557
|
Nat8b
|
N-acetyltransferase 8B |
| chr11_+_38035611 | 0.00 |
ENSRNOT00000087603
ENSRNOT00000002695 |
Mx2
|
MX dynamin like GTPase 2 |
| chr13_-_82758004 | 0.00 |
ENSRNOT00000003932
|
Atp1b1
|
ATPase Na+/K+ transporting subunit beta 1 |
| chr4_+_117371659 | 0.00 |
ENSRNOT00000045735
|
Alms1
|
ALMS1, centrosome and basal body associated protein |
| chr10_+_66509107 | 0.00 |
ENSRNOT00000017230
|
RGD1565317
|
similar to ubiquitin-like/S30 ribosomal fusion protein |
| chr20_+_13827660 | 0.00 |
ENSRNOT00000093131
|
Ddt
|
D-dopachrome tautomerase |
| chr11_-_78341349 | 0.00 |
ENSRNOT00000068116
|
Tp63
|
tumor protein p63 |
| chr6_+_99444013 | 0.00 |
ENSRNOT00000058642
|
Ppp1r36
|
protein phosphatase 1, regulatory subunit 36 |
| chr5_-_78376032 | 0.00 |
ENSRNOT00000075916
|
Alad
|
aminolevulinate dehydratase |
| chr20_-_8019020 | 0.00 |
ENSRNOT00000033119
|
Fkbp5
|
FK506 binding protein 5 |
| chr4_-_164015365 | 0.00 |
ENSRNOT00000078121
|
Ly49si2
|
immunoreceptor Ly49si2 |
| chr10_-_86355006 | 0.00 |
ENSRNOT00000075076
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr1_-_94669461 | 0.00 |
ENSRNOT00000020474
|
Pop4
|
POP4 homolog, ribonuclease P/MRP subunit |
| chr14_+_1462358 | 0.00 |
ENSRNOT00000077243
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr1h2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.0 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.0 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |