Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
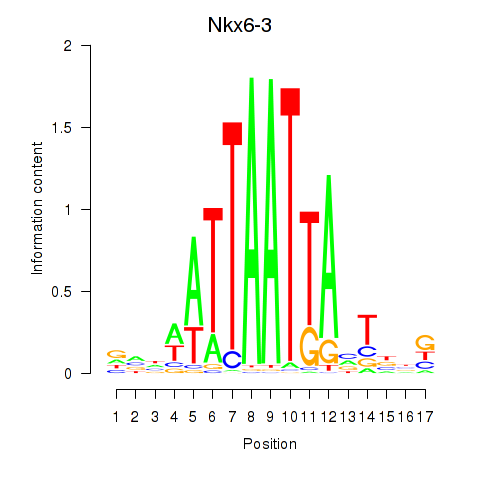
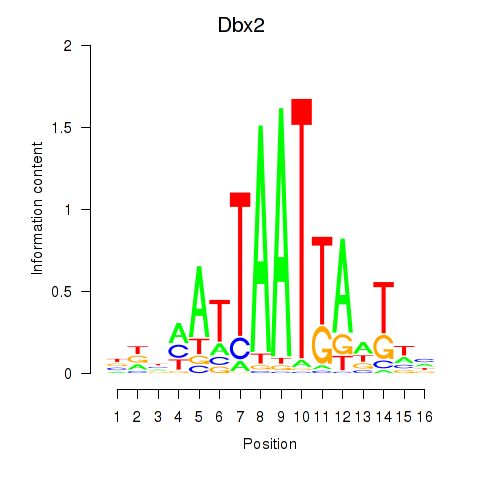
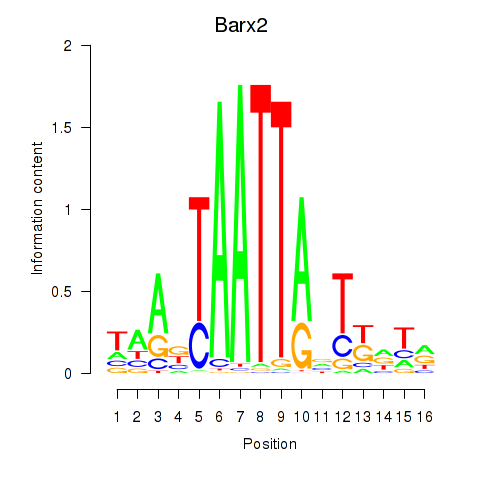
Results for Nkx6-3_Dbx2_Barx2
Z-value: 0.24
Transcription factors associated with Nkx6-3_Dbx2_Barx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-3
|
ENSRNOG00000018147 | NK6 homeobox 3 |
|
Dbx2
|
ENSRNOG00000006885 | developing brain homeobox 2 |
|
Barx2
|
ENSRNOG00000008592 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-3 | rn6_v1_chr16_-_73670482_73670482 | 0.91 | 2.9e-02 | Click! |
| Barx2 | rn6_v1_chr8_-_33017854_33017854 | 0.86 | 6.4e-02 | Click! |
| Dbx2 | rn6_v1_chr7_-_136997050_136997050 | 0.10 | 8.7e-01 | Click! |
Activity profile of Nkx6-3_Dbx2_Barx2 motif
Sorted Z-values of Nkx6-3_Dbx2_Barx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_61685645 | 0.23 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr1_-_190370499 | 0.17 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr9_+_73378057 | 0.12 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr12_-_35979193 | 0.12 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr18_+_14757679 | 0.10 |
ENSRNOT00000071364
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr12_+_25435567 | 0.09 |
ENSRNOT00000092982
|
Gtf2i
|
general transcription factor II I |
| chr1_+_141767940 | 0.08 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr3_+_14889510 | 0.08 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr20_-_13994794 | 0.08 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr1_-_278042312 | 0.08 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chr4_-_66955732 | 0.08 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr18_-_43945273 | 0.07 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr10_+_103206014 | 0.07 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr2_+_122877286 | 0.07 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr2_-_33025271 | 0.07 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr9_+_71915421 | 0.07 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr5_+_22380334 | 0.06 |
ENSRNOT00000009744
|
Clvs1
|
clavesin 1 |
| chr3_+_41019898 | 0.06 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chr14_+_42015347 | 0.06 |
ENSRNOT00000044017
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr2_+_195996521 | 0.06 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr4_-_18396035 | 0.06 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr9_+_73418607 | 0.05 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr13_+_105684420 | 0.05 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr17_-_79085076 | 0.05 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr1_+_140998240 | 0.05 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr2_+_127525285 | 0.05 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr12_-_46493203 | 0.04 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr6_+_64789940 | 0.04 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr10_-_13558377 | 0.04 |
ENSRNOT00000084066
ENSRNOT00000081995 |
Tbc1d24
|
TBC1 domain family, member 24 |
| chr2_-_187786700 | 0.04 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr1_+_167066516 | 0.04 |
ENSRNOT00000000474
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr5_+_135574172 | 0.04 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr1_+_190666149 | 0.04 |
ENSRNOT00000089361
|
LOC102547219
|
uncharacterized LOC102547219 |
| chr3_-_72052990 | 0.04 |
ENSRNOT00000064009
ENSRNOT00000081400 |
Ctnnd1
|
catenin delta 1 |
| chr6_-_3355339 | 0.04 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr1_+_188420461 | 0.03 |
ENSRNOT00000037870
|
Ccp110
|
centriolar coiled-coil protein 110 |
| chr4_+_169161585 | 0.03 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr15_-_93765498 | 0.03 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr12_+_49665282 | 0.03 |
ENSRNOT00000086397
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr3_-_145810834 | 0.03 |
ENSRNOT00000075429
|
LOC102555217
|
zinc finger protein 120-like |
| chr16_+_84465656 | 0.03 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chrX_+_144994139 | 0.03 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr1_+_217345545 | 0.03 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr6_-_4520604 | 0.03 |
ENSRNOT00000042230
ENSRNOT00000043870 ENSRNOT00000070918 ENSRNOT00000046246 ENSRNOT00000052367 ENSRNOT00000042251 |
Slc8a1
|
solute carrier family 8 member A1 |
| chr16_-_10802512 | 0.03 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr14_+_6221085 | 0.03 |
ENSRNOT00000068215
|
AABR07014259.1
|
|
| chr13_-_83457888 | 0.03 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr9_-_100253609 | 0.03 |
ENSRNOT00000036061
|
Kif1a
|
kinesin family member 1A |
| chr2_-_35870578 | 0.03 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chr8_-_69508024 | 0.03 |
ENSRNOT00000083564
|
AABR07070416.3
|
|
| chr3_+_48106099 | 0.03 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr7_-_72328128 | 0.03 |
ENSRNOT00000008227
|
Tspyl5
|
TSPY-like 5 |
| chr15_-_55209342 | 0.02 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr1_-_128287151 | 0.02 |
ENSRNOT00000084946
ENSRNOT00000089723 |
Mef2a
|
myocyte enhancer factor 2a |
| chr8_+_33239139 | 0.02 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr8_-_126390801 | 0.02 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr15_+_37790141 | 0.02 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr14_-_104960141 | 0.02 |
ENSRNOT00000056944
|
Aftph
|
aftiphilin |
| chr17_-_89923423 | 0.02 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr10_-_87286387 | 0.02 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr13_+_109505697 | 0.02 |
ENSRNOT00000005084
|
Angel2
|
angel homolog 2 |
| chr7_-_114590119 | 0.02 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr8_+_32188617 | 0.02 |
ENSRNOT00000081145
|
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr17_+_9596957 | 0.02 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr1_-_241046249 | 0.02 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr12_+_25430464 | 0.02 |
ENSRNOT00000002020
|
Gtf2i
|
general transcription factor II I |
| chr13_+_91054974 | 0.02 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr14_-_82171480 | 0.02 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr1_-_87825333 | 0.02 |
ENSRNOT00000072689
|
Zfp74
|
zinc finger protein 74 |
| chr1_-_90149991 | 0.02 |
ENSRNOT00000076987
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr8_+_82038967 | 0.02 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chrX_+_124321551 | 0.02 |
ENSRNOT00000074486
|
LOC100910807
|
transcriptional regulator Kaiso-like |
| chr4_-_81241152 | 0.02 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr5_+_25725683 | 0.02 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr5_-_16799776 | 0.02 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr7_+_42304534 | 0.02 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr13_+_57243877 | 0.02 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr11_-_82893845 | 0.02 |
ENSRNOT00000075306
|
LOC100912571
|
eukaryotic translation initiation factor 4 gamma 1-like |
| chrX_+_6273733 | 0.02 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr1_-_101819478 | 0.02 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chrX_-_1952111 | 0.02 |
ENSRNOT00000061140
|
Jade3
|
jade family PHD finger 3 |
| chr7_-_139649286 | 0.01 |
ENSRNOT00000080056
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr2_+_257626383 | 0.01 |
ENSRNOT00000080993
|
Zzz3
|
zinc finger, ZZ-type containing 3 |
| chr9_+_66888393 | 0.01 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr14_+_39964588 | 0.01 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr5_-_22765429 | 0.01 |
ENSRNOT00000079432
|
Asph
|
aspartate-beta-hydroxylase |
| chr8_-_39460844 | 0.01 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr1_-_66237501 | 0.01 |
ENSRNOT00000073006
|
Zfp606
|
zinc finger protein 606 |
| chr19_-_43955747 | 0.01 |
ENSRNOT00000058891
ENSRNOT00000041815 |
Bcar1
|
BCAR1, Cas family scaffolding protein |
| chr20_+_3155652 | 0.01 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr2_-_198706428 | 0.01 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr5_-_56567320 | 0.01 |
ENSRNOT00000066081
|
Topors
|
TOP1 binding arginine/serine rich protein |
| chr7_-_140291620 | 0.01 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr2_-_60657712 | 0.01 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr10_-_52290657 | 0.01 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr3_+_8430829 | 0.01 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr15_+_62406873 | 0.01 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr5_+_34007920 | 0.01 |
ENSRNOT00000009611
|
Ttpa
|
alpha tocopherol transfer protein |
| chr2_+_123597340 | 0.01 |
ENSRNOT00000058552
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr14_-_7824209 | 0.01 |
ENSRNOT00000087081
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr1_+_105285419 | 0.01 |
ENSRNOT00000089693
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr2_+_220298245 | 0.01 |
ENSRNOT00000022625
|
Plppr4
|
phospholipid phosphatase related 4 |
| chr11_+_31539016 | 0.01 |
ENSRNOT00000072856
|
Ifnar2
|
interferon alpha and beta receptor subunit 2 |
| chr9_+_71247781 | 0.01 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr1_+_101599018 | 0.01 |
ENSRNOT00000028494
|
Fut1
|
fucosyltransferase 1 |
| chr3_-_15278645 | 0.01 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr6_-_3254779 | 0.01 |
ENSRNOT00000061975
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr4_+_32541706 | 0.01 |
ENSRNOT00000050827
|
Sdhaf3
|
succinate dehydrogenase complex assembly factor 3 |
| chr8_+_73682887 | 0.01 |
ENSRNOT00000057522
|
Vps13c
|
vacuolar protein sorting 13C |
| chr16_-_20562399 | 0.01 |
ENSRNOT00000031692
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr2_-_149444548 | 0.01 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr16_+_6048004 | 0.01 |
ENSRNOT00000020750
|
Actr8
|
ARP8 actin-related protein 8 homolog |
| chr2_-_117454769 | 0.01 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr1_-_107373807 | 0.01 |
ENSRNOT00000056024
|
Svip
|
small VCP interacting protein |
| chr1_-_266428239 | 0.01 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chrX_+_9956370 | 0.01 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr8_-_118436894 | 0.01 |
ENSRNOT00000073395
|
LOC100911807
|
ATR-interacting protein-like |
| chr2_+_248398917 | 0.01 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr3_+_129885826 | 0.01 |
ENSRNOT00000009743
|
Slx4ip
|
SLX4 interacting protein |
| chr9_+_50966766 | 0.01 |
ENSRNOT00000076636
|
Ercc5
|
ERCC excision repair 5, endonuclease |
| chr10_+_67810810 | 0.01 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr12_-_23727535 | 0.01 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr5_+_28485619 | 0.01 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr4_-_168656673 | 0.01 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr10_-_16752205 | 0.01 |
ENSRNOT00000079462
|
Crebrf
|
CREB3 regulatory factor |
| chr13_+_56546021 | 0.01 |
ENSRNOT00000016797
|
Aspm
|
abnormal spindle microtubule assembly |
| chr1_+_1545134 | 0.01 |
ENSRNOT00000044523
|
AABR07000156.1
|
|
| chr19_-_19727081 | 0.01 |
ENSRNOT00000020700
|
Adcy7
|
adenylate cyclase 7 |
| chr5_+_25253010 | 0.01 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr5_+_58995249 | 0.00 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr16_+_71738718 | 0.00 |
ENSRNOT00000022097
|
Plekha2
|
pleckstrin homology domain containing A2 |
| chr3_-_66394764 | 0.00 |
ENSRNOT00000075832
|
Cerkl
|
ceramide kinase-like |
| chr2_+_220432037 | 0.00 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr8_-_49308806 | 0.00 |
ENSRNOT00000047291
|
Cd3e
|
CD3e molecule |
| chr13_-_90405591 | 0.00 |
ENSRNOT00000006849
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chr10_+_29033989 | 0.00 |
ENSRNOT00000088322
|
Slu7
|
SLU7 homolog, splicing factor |
| chr2_+_86996497 | 0.00 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr18_-_16543992 | 0.00 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chrX_+_84064427 | 0.00 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr3_+_17889972 | 0.00 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr14_-_81053905 | 0.00 |
ENSRNOT00000045068
ENSRNOT00000040215 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr4_+_68635581 | 0.00 |
ENSRNOT00000016217
|
Ssbp1
|
single stranded DNA binding protein 1 |
| chr13_+_113373578 | 0.00 |
ENSRNOT00000009900
|
Plxna2
|
plexin A2 |
| chr1_-_228014924 | 0.00 |
ENSRNOT00000042091
|
Oosp2
|
oocyte secreted protein 2 |
| chr3_-_57607683 | 0.00 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr1_+_68436917 | 0.00 |
ENSRNOT00000088586
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
| chr14_-_33444235 | 0.00 |
ENSRNOT00000044441
|
Arl9
|
ADP-ribosylation factor like GTPase 9 |
| chr3_+_55910177 | 0.00 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr15_-_80713153 | 0.00 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr16_+_24978527 | 0.00 |
ENSRNOT00000019363
|
Tma16
|
translation machinery associated 16 homolog |
| chr7_+_44009069 | 0.00 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr4_+_117962319 | 0.00 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr1_+_68436593 | 0.00 |
ENSRNOT00000080325
|
LOC108348118
|
leucyl-cystinyl aminopeptidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-3_Dbx2_Barx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.0 | GO:0043474 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.0 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |