Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Nkx6-2
Z-value: 0.23
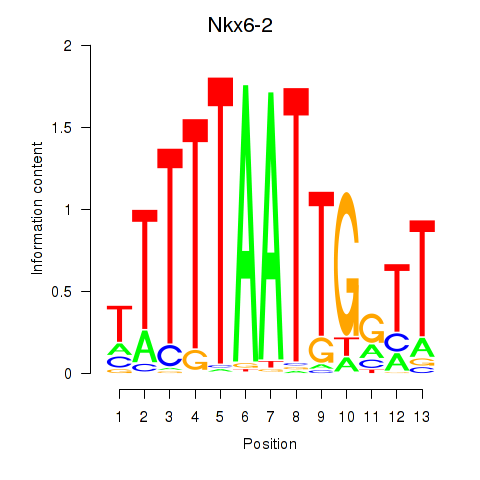
Transcription factors associated with Nkx6-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-2
|
ENSRNOG00000017748 | NK6 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-2 | rn6_v1_chr1_-_211923929_211923929 | -0.69 | 2.0e-01 | Click! |
Activity profile of Nkx6-2 motif
Sorted Z-values of Nkx6-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_21970103 | 0.08 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chr14_+_104250617 | 0.07 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr19_+_27603307 | 0.07 |
ENSRNOT00000074702
|
AABR07043347.1
|
|
| chr10_-_104482838 | 0.06 |
ENSRNOT00000007246
|
Recql5
|
RecQ like helicase 5 |
| chr3_+_113918629 | 0.06 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr6_-_76552559 | 0.05 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr15_+_87722221 | 0.05 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr18_+_1723565 | 0.05 |
ENSRNOT00000033468
|
Greb1l
|
growth regulation by estrogen in breast cancer 1 like |
| chr10_+_65552897 | 0.04 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr5_+_173447784 | 0.04 |
ENSRNOT00000027251
|
Tnfrsf4
|
TNF receptor superfamily member 4 |
| chr8_+_23398030 | 0.04 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr7_-_122644054 | 0.04 |
ENSRNOT00000025941
|
Dnajb7
|
DnaJ heat shock protein family (Hsp40) member B7 |
| chr2_+_210045161 | 0.04 |
ENSRNOT00000024455
|
Slc16a4
|
solute carrier family 16, member 4 |
| chr11_-_62451149 | 0.04 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_64147251 | 0.04 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr2_-_96509424 | 0.03 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr18_+_30904498 | 0.03 |
ENSRNOT00000026969
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr2_-_188559882 | 0.03 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr6_+_86872903 | 0.03 |
ENSRNOT00000086894
|
Fancm
|
Fanconi anemia, complementation group M |
| chr9_+_73378057 | 0.03 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr3_+_112390106 | 0.03 |
ENSRNOT00000081197
|
LOC691918
|
similar to Centrosomal protein of 27 kDa (Cep27 protein) |
| chr13_-_111972603 | 0.03 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr8_+_115546893 | 0.03 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr3_-_7796385 | 0.03 |
ENSRNOT00000082489
|
Ntng2
|
netrin G2 |
| chr16_-_10802512 | 0.03 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr4_-_150522351 | 0.02 |
ENSRNOT00000079053
|
Zfp9
|
zinc finger protein 9 |
| chr2_+_198726118 | 0.02 |
ENSRNOT00000056237
|
Lix1l
|
limb and CNS expressed 1 like |
| chr3_+_48082935 | 0.02 |
ENSRNOT00000087711
ENSRNOT00000067545 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr3_+_56030915 | 0.02 |
ENSRNOT00000010489
|
Phospho2
|
phosphatase, orphan 2 |
| chr4_+_88694583 | 0.02 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr7_-_11223649 | 0.02 |
ENSRNOT00000061191
|
Mfsd12
|
major facilitator superfamily domain containing 12 |
| chr13_-_52413681 | 0.02 |
ENSRNOT00000037100
ENSRNOT00000011801 |
Nav1
|
neuron navigator 1 |
| chr18_+_29124428 | 0.02 |
ENSRNOT00000073131
|
Igip
|
IgA-inducing protein |
| chr10_-_43599232 | 0.02 |
ENSRNOT00000044299
|
Gemin5
|
gem (nuclear organelle) associated protein 5 |
| chr17_+_35435079 | 0.02 |
ENSRNOT00000074800
|
Exoc2
|
exocyst complex component 2 |
| chr14_+_63095720 | 0.02 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr13_-_104284068 | 0.02 |
ENSRNOT00000005407
|
LOC100912365
|
uncharacterized LOC100912365 |
| chr7_-_68512397 | 0.02 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr13_-_47916185 | 0.02 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr17_-_53915076 | 0.02 |
ENSRNOT00000010836
|
Arid4b
|
AT-rich interaction domain 4B |
| chr3_-_48831417 | 0.02 |
ENSRNOT00000009920
ENSRNOT00000085246 |
Kcnh7
|
potassium voltage-gated channel subfamily H member 7 |
| chr6_-_98606109 | 0.02 |
ENSRNOT00000006860
|
Wdr89
|
WD repeat domain 89 |
| chr5_+_4373626 | 0.02 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr9_+_1313209 | 0.02 |
ENSRNOT00000083387
|
Tbc1d5
|
TBC1 domain family, member 5 |
| chr17_+_9596957 | 0.02 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr4_-_135069970 | 0.02 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr5_-_166116516 | 0.02 |
ENSRNOT00000079919
ENSRNOT00000080888 |
Kif1b
|
kinesin family member 1B |
| chr8_+_85503224 | 0.02 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr17_+_34704616 | 0.02 |
ENSRNOT00000090706
ENSRNOT00000083674 ENSRNOT00000077110 |
Exoc2
|
exocyst complex component 2 |
| chr18_-_77535564 | 0.02 |
ENSRNOT00000041380
ENSRNOT00000090265 |
Atp9b
|
ATPase phospholipid transporting 9B (putative) |
| chr16_+_46731403 | 0.02 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr6_+_129621070 | 0.02 |
ENSRNOT00000091338
|
Papola
|
poly (A) polymerase alpha |
| chr9_+_53546018 | 0.01 |
ENSRNOT00000016842
|
Inpp1
|
inositol polyphosphate-1-phosphatase |
| chr4_-_51931112 | 0.01 |
ENSRNOT00000080575
|
Pot1
|
protection of telomeres 1 |
| chr18_-_26211445 | 0.01 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr15_+_56666012 | 0.01 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr3_-_56030744 | 0.01 |
ENSRNOT00000089637
|
Ccdc173
|
coiled-coil domain containing 173 |
| chrX_+_22342308 | 0.01 |
ENSRNOT00000079909
ENSRNOT00000087945 ENSRNOT00000077528 |
Kdm5c
|
lysine demethylase 5C |
| chr7_+_64769089 | 0.01 |
ENSRNOT00000088861
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr4_+_162281574 | 0.01 |
ENSRNOT00000087850
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr18_+_30496318 | 0.01 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr3_+_8430829 | 0.01 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chrX_-_25590048 | 0.01 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr14_+_34455934 | 0.01 |
ENSRNOT00000085991
ENSRNOT00000002981 |
Clock
|
clock circadian regulator |
| chr1_+_100577056 | 0.01 |
ENSRNOT00000026992
|
Napsa
|
napsin A aspartic peptidase |
| chr16_-_10661528 | 0.01 |
ENSRNOT00000085012
|
Fam35a
|
family with sequence similarity 35, member A |
| chr14_-_72025137 | 0.01 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr13_+_80464348 | 0.01 |
ENSRNOT00000076324
|
Vamp4
|
vesicle-associated membrane protein 4 |
| chr8_-_109621408 | 0.01 |
ENSRNOT00000087398
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr2_+_92559929 | 0.01 |
ENSRNOT00000033404
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr16_+_32449116 | 0.01 |
ENSRNOT00000050549
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr11_-_25456836 | 0.01 |
ENSRNOT00000085444
|
Adamts5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr3_+_152294656 | 0.01 |
ENSRNOT00000027067
|
Phf20
|
PHD finger protein 20 |
| chr12_+_41316764 | 0.01 |
ENSRNOT00000090867
|
Oas3
|
2'-5'-oligoadenylate synthetase 3 |
| chr7_-_132143470 | 0.01 |
ENSRNOT00000038946
ENSRNOT00000044092 ENSRNOT00000066528 ENSRNOT00000045553 ENSRNOT00000046744 ENSRNOT00000055742 |
Kif21a
|
kinesin family member 21A |
| chr5_+_133819726 | 0.01 |
ENSRNOT00000081075
|
Stil
|
Scl/Tal1 interrupting locus |
| chr1_-_251387002 | 0.01 |
ENSRNOT00000092063
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr20_+_41266566 | 0.01 |
ENSRNOT00000000653
|
Frk
|
fyn-related Src family tyrosine kinase |
| chr14_-_72122158 | 0.01 |
ENSRNOT00000064495
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chrX_-_63291371 | 0.01 |
ENSRNOT00000082421
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr3_-_113423473 | 0.01 |
ENSRNOT00000064982
|
Serinc4
|
serine incorporator 4 |
| chr3_-_104018861 | 0.01 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr20_+_49319307 | 0.01 |
ENSRNOT00000057078
|
Atg5
|
autophagy related 5 |
| chr7_-_40315377 | 0.01 |
ENSRNOT00000091319
ENSRNOT00000036141 |
RGD1307947
|
similar to RIKEN cDNA C430008C19 |
| chr2_+_127459012 | 0.01 |
ENSRNOT00000093685
ENSRNOT00000072675 |
Intu
|
inturned planar cell polarity protein |
| chr12_-_51877624 | 0.01 |
ENSRNOT00000056800
|
Chek2
|
checkpoint kinase 2 |
| chr19_-_37528011 | 0.01 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr2_-_128675408 | 0.01 |
ENSRNOT00000019256
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr10_+_38661901 | 0.01 |
ENSRNOT00000009098
|
Zcchc10
|
zinc finger CCHC-type containing 10 |
| chr6_-_38007162 | 0.01 |
ENSRNOT00000044583
|
AABR07063632.1
|
|
| chr3_-_48604097 | 0.01 |
ENSRNOT00000009620
|
Ifih1
|
interferon induced with helicase C domain 1 |
| chrX_+_124631881 | 0.01 |
ENSRNOT00000009329
|
Atp1b4
|
ATPase Na+/K+ transporting family member beta 4 |
| chr9_+_73334618 | 0.01 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr8_+_85259982 | 0.01 |
ENSRNOT00000010409
|
Elovl5
|
ELOVL fatty acid elongase 5 |
| chr6_+_108009251 | 0.01 |
ENSRNOT00000059073
|
Ptgr2
|
prostaglandin reductase 2 |
| chr15_-_88036354 | 0.01 |
ENSRNOT00000014747
|
Ednrb
|
endothelin receptor type B |
| chrX_+_110016995 | 0.00 |
ENSRNOT00000093542
|
Nrk
|
Nik related kinase |
| chrX_+_157759624 | 0.00 |
ENSRNOT00000003192
|
LOC686087
|
similar to motile sperm domain containing 1 |
| chr19_-_11669578 | 0.00 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr4_-_66955732 | 0.00 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr1_-_67284864 | 0.00 |
ENSRNOT00000082908
|
LOC691661
|
similar to zinc finger and SCAN domain containing 4 |
| chrX_-_138237157 | 0.00 |
ENSRNOT00000003414
|
Rap2c
|
RAP2C, member of RAS oncogene family |
| chr6_-_95934296 | 0.00 |
ENSRNOT00000034338
|
Six1
|
SIX homeobox 1 |
| chr13_+_67545430 | 0.00 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr11_-_44292884 | 0.00 |
ENSRNOT00000084060
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr3_+_55960067 | 0.00 |
ENSRNOT00000010216
|
Ppig
|
peptidylprolyl isomerase G |
| chr3_-_113423149 | 0.00 |
ENSRNOT00000021008
|
Serinc4
|
serine incorporator 4 |
| chr16_-_930527 | 0.00 |
ENSRNOT00000014787
|
Spin1
|
spindlin 1 |
| chr10_+_104483042 | 0.00 |
ENSRNOT00000007362
|
Sap30bp
|
SAP30 binding protein |
| chr2_+_41442241 | 0.00 |
ENSRNOT00000067546
|
Pde4d
|
phosphodiesterase 4D |
| chr10_+_63635219 | 0.00 |
ENSRNOT00000081813
ENSRNOT00000005016 |
Prpf8
|
pre-mRNA processing factor 8 |
| chr8_+_85355766 | 0.00 |
ENSRNOT00000010583
|
Gcm1
|
glial cells missing homolog 1 |
| chr10_+_56445647 | 0.00 |
ENSRNOT00000056870
|
Tmem256
|
transmembrane protein 256 |
| chr2_+_128675814 | 0.00 |
ENSRNOT00000058366
|
RGD1359508
|
similar to protein C33A12.3 |
| chr4_-_163762434 | 0.00 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr8_+_116657371 | 0.00 |
ENSRNOT00000024966
|
Mon1a
|
MON1 homolog A, secretory trafficking associated |
| chr3_-_60611924 | 0.00 |
ENSRNOT00000068745
|
Chn1
|
chimerin 1 |
| chr5_-_102743417 | 0.00 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr11_-_71757801 | 0.00 |
ENSRNOT00000029572
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_-_231530690 | 0.00 |
ENSRNOT00000087140
ENSRNOT00000018452 ENSRNOT00000018272 |
Tle4
|
transducin-like enhancer of split 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |