Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
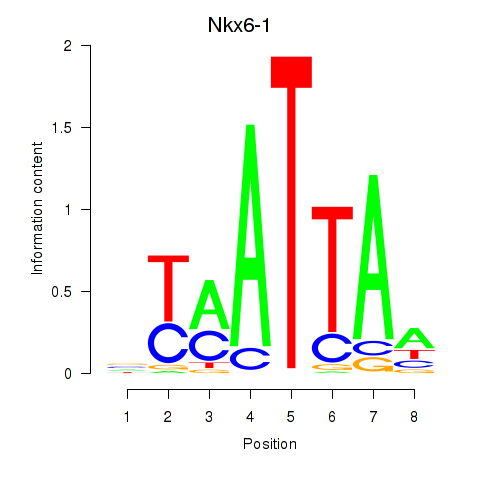
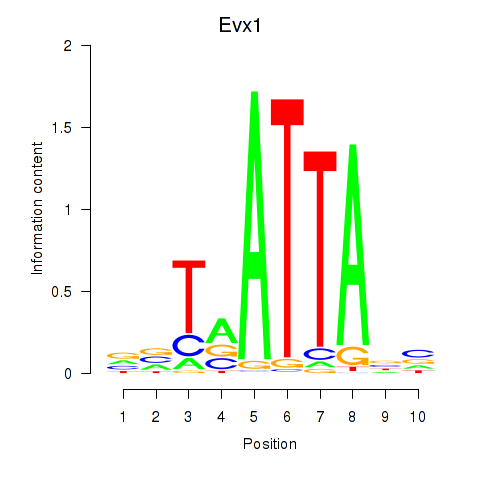
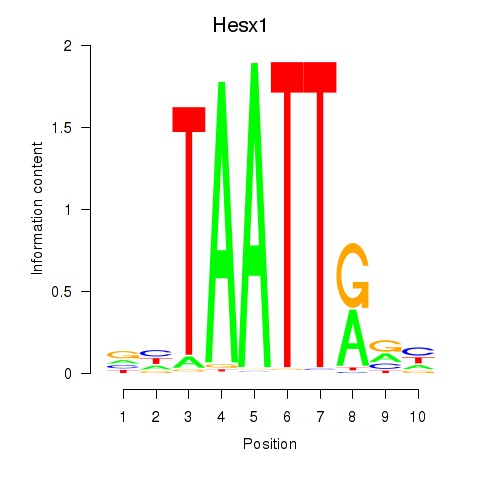
Results for Nkx6-1_Evx1_Hesx1
Z-value: 0.39
Transcription factors associated with Nkx6-1_Evx1_Hesx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx6-1
|
ENSRNOG00000002149 | NK6 homeobox 1 |
|
Hesx1
|
ENSRNOG00000014133 | HESX homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx6-1 | rn6_v1_chr14_+_9555264_9555264 | 0.56 | 3.3e-01 | Click! |
Activity profile of Nkx6-1_Evx1_Hesx1 motif
Sorted Z-values of Nkx6-1_Evx1_Hesx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 1.61 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr3_-_66417741 | 0.81 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr2_+_145174876 | 0.53 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr12_-_17186679 | 0.50 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr4_+_94696965 | 0.36 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr4_+_86275717 | 0.25 |
ENSRNOT00000016414
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr17_+_11683862 | 0.24 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr1_-_124803363 | 0.20 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr3_+_95715193 | 0.20 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr12_-_45801842 | 0.18 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr6_-_125723732 | 0.17 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr16_-_59366824 | 0.15 |
ENSRNOT00000015062
|
RGD1304810
|
similar to 6430573F11Rik protein |
| chr6_+_58468155 | 0.15 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chrX_+_111735820 | 0.14 |
ENSRNOT00000086948
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr6_-_106971250 | 0.14 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr19_+_25043680 | 0.13 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr3_-_101547478 | 0.13 |
ENSRNOT00000006203
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr6_-_115616766 | 0.13 |
ENSRNOT00000006143
ENSRNOT00000045870 |
Sel1l
|
SEL1L ERAD E3 ligase adaptor subunit |
| chr1_+_13595295 | 0.13 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr1_-_190370499 | 0.12 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chrX_+_74200972 | 0.12 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr13_+_90533365 | 0.12 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr17_+_63635086 | 0.12 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr1_+_217345545 | 0.12 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr10_-_91986632 | 0.11 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr10_+_61685645 | 0.11 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr1_+_264504591 | 0.11 |
ENSRNOT00000050076
|
Pax2
|
paired box 2 |
| chr1_-_215033460 | 0.10 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr1_-_67065797 | 0.10 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr4_+_68656928 | 0.10 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr5_-_136965191 | 0.10 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr10_-_87286387 | 0.10 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chrX_-_124464963 | 0.10 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr9_+_71915421 | 0.09 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr3_-_141411170 | 0.09 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr3_+_14889510 | 0.09 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr18_+_30435119 | 0.09 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr5_-_150167077 | 0.08 |
ENSRNOT00000088493
ENSRNOT00000013761 ENSRNOT00000067349 ENSRNOT00000051097 ENSRNOT00000067189 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr1_-_90149991 | 0.08 |
ENSRNOT00000076987
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr18_-_26656879 | 0.08 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr16_+_54319377 | 0.08 |
ENSRNOT00000090266
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr8_-_78233430 | 0.08 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr9_+_117795132 | 0.08 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chr2_-_33025271 | 0.08 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_144052061 | 0.08 |
ENSRNOT00000020103
ENSRNOT00000091378 |
Amhr2
|
anti-Mullerian hormone receptor type 2 |
| chr6_+_101532518 | 0.07 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr3_-_7498555 | 0.07 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr18_+_30592794 | 0.07 |
ENSRNOT00000027133
|
Pcdhb22
|
protocadherin beta 22 |
| chr12_-_35979193 | 0.07 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr18_-_31343797 | 0.07 |
ENSRNOT00000081850
|
Pcdh1
|
protocadherin 1 |
| chr3_+_48106099 | 0.07 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr10_-_74679858 | 0.07 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr10_-_88670430 | 0.07 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr1_+_87224677 | 0.07 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr4_+_158088505 | 0.07 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr4_+_117962319 | 0.07 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr19_-_17399548 | 0.06 |
ENSRNOT00000075991
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr6_+_43234526 | 0.06 |
ENSRNOT00000086808
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
| chr2_-_231409988 | 0.06 |
ENSRNOT00000080637
ENSRNOT00000037255 ENSRNOT00000074002 |
Ank2
|
ankyrin 2 |
| chr2_-_96509424 | 0.06 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr1_-_43638161 | 0.06 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr10_-_40953467 | 0.06 |
ENSRNOT00000092189
|
Glra1
|
glycine receptor, alpha 1 |
| chr4_+_22898527 | 0.06 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr1_-_275882444 | 0.06 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr12_-_10335499 | 0.06 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr1_-_201906286 | 0.06 |
ENSRNOT00000064511
ENSRNOT00000036625 |
RGD1560958
|
similar to RIKEN cDNA 1700063I17 |
| chr16_+_54332660 | 0.06 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr9_+_73418607 | 0.06 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr5_+_135574172 | 0.06 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr19_-_37528011 | 0.06 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr1_+_190666149 | 0.06 |
ENSRNOT00000089361
|
LOC102547219
|
uncharacterized LOC102547219 |
| chr1_+_105284753 | 0.06 |
ENSRNOT00000041950
|
Slc6a5
|
solute carrier family 6 member 5 |
| chr2_-_32518643 | 0.06 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_136182224 | 0.05 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr18_+_27558089 | 0.05 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chr3_+_54253949 | 0.05 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr10_-_84920886 | 0.05 |
ENSRNOT00000068083
|
Sp2
|
Sp2 transcription factor |
| chrM_+_7758 | 0.05 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr9_+_10941613 | 0.05 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr1_+_240355149 | 0.05 |
ENSRNOT00000018521
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr13_-_73056765 | 0.05 |
ENSRNOT00000000049
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr1_+_40879747 | 0.05 |
ENSRNOT00000083702
|
Akap12
|
A-kinase anchoring protein 12 |
| chr3_-_57607683 | 0.05 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr4_+_88694583 | 0.05 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr5_-_12526962 | 0.05 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr3_+_56125924 | 0.05 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr1_+_277355619 | 0.05 |
ENSRNOT00000022788
|
Nhlrc2
|
NHL repeat containing 2 |
| chr4_+_169161585 | 0.05 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr1_-_67390141 | 0.05 |
ENSRNOT00000025808
|
Sbk1
|
SH3 domain binding kinase 1 |
| chrX_+_6273733 | 0.05 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr1_-_278042312 | 0.05 |
ENSRNOT00000018693
|
Ablim1
|
actin-binding LIM protein 1 |
| chr3_+_128828331 | 0.05 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chrX_+_65040934 | 0.05 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chrX_+_118197217 | 0.05 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr10_+_103206014 | 0.05 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr4_-_168517177 | 0.05 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr18_-_32670665 | 0.05 |
ENSRNOT00000019409
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr10_+_66942398 | 0.05 |
ENSRNOT00000018986
|
Rab11fip4
|
RAB11 family interacting protein 4 |
| chr15_-_26678420 | 0.05 |
ENSRNOT00000041420
|
RGD1310110
|
similar to 3632451O06Rik protein |
| chr15_+_11298478 | 0.05 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr2_+_217963456 | 0.04 |
ENSRNOT00000024243
|
Olfm3
|
olfactomedin 3 |
| chr10_+_91217079 | 0.04 |
ENSRNOT00000004218
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr11_-_44292884 | 0.04 |
ENSRNOT00000084060
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr10_-_67285617 | 0.04 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr5_-_168734296 | 0.04 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr8_+_23193181 | 0.04 |
ENSRNOT00000071703
|
Zfp872
|
zinc finger protein 872 |
| chr14_+_45982244 | 0.04 |
ENSRNOT00000076881
ENSRNOT00000002984 |
Rell1
|
RELT-like 1 |
| chr19_-_37938857 | 0.04 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr11_+_57207656 | 0.04 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr10_-_13446135 | 0.04 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr20_+_6211420 | 0.04 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr4_+_180062799 | 0.04 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr17_-_43776460 | 0.04 |
ENSRNOT00000089055
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr5_+_139394794 | 0.04 |
ENSRNOT00000045954
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr6_+_104291071 | 0.04 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr3_+_148654668 | 0.04 |
ENSRNOT00000081370
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr2_+_185846232 | 0.04 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr13_+_87986240 | 0.04 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr3_+_151335292 | 0.04 |
ENSRNOT00000073642
|
Mmp24
|
matrix metallopeptidase 24 |
| chr20_+_27975549 | 0.04 |
ENSRNOT00000092075
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr2_-_187786700 | 0.04 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chrX_+_74205842 | 0.04 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr3_+_45683993 | 0.04 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr12_-_19167015 | 0.04 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr1_+_204959174 | 0.04 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr5_-_65073012 | 0.04 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr3_-_138683318 | 0.04 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr7_-_59547174 | 0.04 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr10_-_87232723 | 0.04 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr2_+_266315036 | 0.04 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr3_-_161917285 | 0.04 |
ENSRNOT00000025150
|
Cdh22
|
cadherin 22 |
| chr4_-_18396035 | 0.04 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr1_-_175657485 | 0.03 |
ENSRNOT00000024561
|
rnf141
|
ring finger protein 141 |
| chr6_-_125723944 | 0.03 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr7_-_15856676 | 0.03 |
ENSRNOT00000086553
|
AABR07055919.1
|
|
| chr14_-_114692764 | 0.03 |
ENSRNOT00000008210
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr4_-_6978073 | 0.03 |
ENSRNOT00000045311
|
Nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr12_+_17253791 | 0.03 |
ENSRNOT00000083814
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr2_-_188559882 | 0.03 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr17_-_22678253 | 0.03 |
ENSRNOT00000083122
|
LOC100362172
|
LRRGT00112-like |
| chr16_-_79700992 | 0.03 |
ENSRNOT00000016420
|
Kbtbd11
|
kelch repeat and BTB domain containing 11 |
| chr7_+_141666111 | 0.03 |
ENSRNOT00000083250
|
AABR07058886.1
|
|
| chr4_-_130659697 | 0.03 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr2_-_123972356 | 0.03 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr14_+_76866135 | 0.03 |
ENSRNOT00000037291
|
Zfp518b
|
zinc finger protein 518B |
| chr16_+_84465656 | 0.03 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr5_-_2982603 | 0.03 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr19_+_38039564 | 0.03 |
ENSRNOT00000087491
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chr6_-_107325345 | 0.03 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr12_+_47179664 | 0.03 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr1_+_140998240 | 0.03 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr7_+_11414446 | 0.03 |
ENSRNOT00000027441
|
Pias4
|
protein inhibitor of activated STAT, 4 |
| chr6_+_28382962 | 0.03 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr2_+_226563050 | 0.03 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr8_-_63750531 | 0.03 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr5_+_131380297 | 0.03 |
ENSRNOT00000010548
|
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr3_-_148057523 | 0.03 |
ENSRNOT00000055408
|
Defb24
|
defensin beta 24 |
| chr18_+_14757679 | 0.03 |
ENSRNOT00000071364
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr18_+_14471213 | 0.03 |
ENSRNOT00000045224
|
Dtna
|
dystrobrevin, alpha |
| chr2_+_207930796 | 0.03 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr19_-_58735173 | 0.03 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr9_-_65736852 | 0.03 |
ENSRNOT00000087458
|
Trak2
|
trafficking kinesin protein 2 |
| chr5_+_187312 | 0.03 |
ENSRNOT00000030857
|
LOC102557117
|
zinc finger protein 120-like |
| chr10_-_87136026 | 0.03 |
ENSRNOT00000014230
ENSRNOT00000083233 |
Smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr2_-_231409496 | 0.03 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chr3_+_38559914 | 0.03 |
ENSRNOT00000085545
|
Fmnl2
|
formin-like 2 |
| chr3_-_11452529 | 0.03 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chr18_+_1723565 | 0.03 |
ENSRNOT00000033468
|
Greb1l
|
growth regulation by estrogen in breast cancer 1 like |
| chr14_-_44078897 | 0.03 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr5_-_22765429 | 0.03 |
ENSRNOT00000079432
|
Asph
|
aspartate-beta-hydroxylase |
| chr14_-_115052450 | 0.03 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr14_+_34446616 | 0.03 |
ENSRNOT00000002976
|
Clock
|
clock circadian regulator |
| chr14_-_14192048 | 0.03 |
ENSRNOT00000060868
|
Bmp2k
|
BMP2 inducible kinase |
| chr1_-_198450047 | 0.03 |
ENSRNOT00000027360
|
Mvp
|
major vault protein |
| chr3_+_48096954 | 0.03 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr6_+_8219385 | 0.03 |
ENSRNOT00000040509
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr13_+_31081804 | 0.03 |
ENSRNOT00000041413
|
Cdh7
|
cadherin 7 |
| chr18_-_57866741 | 0.03 |
ENSRNOT00000025790
|
Fbxo38
|
F-box protein 38 |
| chr1_-_189182306 | 0.03 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr6_-_44363915 | 0.03 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr20_+_3558827 | 0.03 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_14091766 | 0.03 |
ENSRNOT00000075373
|
AABR07027088.1
|
|
| chr13_+_49005405 | 0.03 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr4_-_68597586 | 0.03 |
ENSRNOT00000015921
|
RGD1563986
|
similar to RIKEN cDNA E330009J07 gene |
| chr4_-_51199570 | 0.03 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr4_+_172119331 | 0.03 |
ENSRNOT00000010579
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr17_-_89923423 | 0.03 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr3_-_103745236 | 0.03 |
ENSRNOT00000006876
|
Nutm1
|
NUT midline carcinoma, family member 1 |
| chr6_+_2216623 | 0.03 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr4_-_98305173 | 0.03 |
ENSRNOT00000010151
|
Il23r
|
interleukin 23 receptor |
| chr6_-_98666007 | 0.03 |
ENSRNOT00000082695
|
AABR07064867.1
|
|
| chr5_+_58855773 | 0.03 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr14_+_104250617 | 0.03 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr1_+_80279706 | 0.03 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr5_+_22392732 | 0.03 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr1_-_197568165 | 0.03 |
ENSRNOT00000079044
|
Xpo6
|
exportin 6 |
| chr12_-_52658275 | 0.03 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr12_+_24761210 | 0.03 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr6_-_3355339 | 0.03 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr10_+_35870232 | 0.03 |
ENSRNOT00000078505
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx6-1_Evx1_Hesx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 0.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.2 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 0.5 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.0 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0070668 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.0 | GO:1990168 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.0 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |