Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
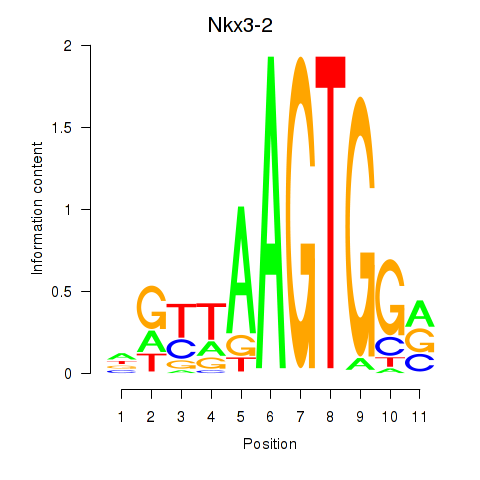
Results for Nkx3-2
Z-value: 0.60
Transcription factors associated with Nkx3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx3-2
|
ENSRNOG00000060782 | NK3 homeobox 2 |
Activity profile of Nkx3-2 motif
Sorted Z-values of Nkx3-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_260596777 | 0.43 |
ENSRNOT00000009370
|
Lhx8
|
LIM homeobox 8 |
| chr6_+_8886591 | 0.41 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr14_-_37770059 | 0.33 |
ENSRNOT00000029721
|
Slc10a4
|
solute carrier family 10, member 4 |
| chr10_-_15577977 | 0.31 |
ENSRNOT00000052292
|
Hba-a3
|
hemoglobin alpha, adult chain 3 |
| chr5_-_17061361 | 0.25 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr8_+_48569328 | 0.25 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr10_+_110138586 | 0.24 |
ENSRNOT00000086096
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr5_-_17061837 | 0.23 |
ENSRNOT00000011892
|
Penk
|
proenkephalin |
| chr1_+_79894450 | 0.21 |
ENSRNOT00000057963
|
Nanos2
|
nanos C2HC-type zinc finger 2 |
| chr9_+_20004280 | 0.21 |
ENSRNOT00000075670
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr10_-_27862868 | 0.18 |
ENSRNOT00000004877
|
Gabra6
|
gamma-aminobutyric acid type A receptor alpha 6 subunit |
| chr8_-_46694613 | 0.18 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr12_-_22472044 | 0.15 |
ENSRNOT00000073356
|
Ufsp1
|
UFM1-specific peptidase 1 |
| chr18_+_79406381 | 0.14 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr10_+_74874130 | 0.14 |
ENSRNOT00000076050
ENSRNOT00000044226 ENSRNOT00000085052 ENSRNOT00000076196 |
Sept4
|
septin 4 |
| chr9_-_78969013 | 0.14 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr4_-_30338679 | 0.13 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr7_+_116632506 | 0.13 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr1_-_7801438 | 0.13 |
ENSRNOT00000022273
|
AABR07000276.1
|
|
| chr11_-_72109964 | 0.13 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr7_-_107775712 | 0.12 |
ENSRNOT00000010811
ENSRNOT00000093459 |
Ndrg1
|
N-myc downstream regulated 1 |
| chr5_+_129257429 | 0.12 |
ENSRNOT00000072396
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr9_+_67763897 | 0.11 |
ENSRNOT00000071226
|
Icos
|
inducible T-cell co-stimulator |
| chr8_+_128824508 | 0.11 |
ENSRNOT00000025343
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr4_-_70996395 | 0.10 |
ENSRNOT00000021485
|
Kel
|
Kell blood group, metallo-endopeptidase |
| chr1_-_89068348 | 0.10 |
ENSRNOT00000033621
|
Upk1a
|
uroplakin 1A |
| chr11_+_70687500 | 0.10 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr15_+_4475051 | 0.10 |
ENSRNOT00000008461
|
Kcnk16
|
potassium two pore domain channel subfamily K member 16 |
| chr1_-_141481315 | 0.09 |
ENSRNOT00000020229
|
Pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr5_-_138222534 | 0.09 |
ENSRNOT00000009691
|
Zfp691
|
zinc finger protein 691 |
| chr10_-_56558487 | 0.09 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr12_-_13924531 | 0.09 |
ENSRNOT00000001477
|
Slc29a4
|
solute carrier family 29 member 4 |
| chr12_+_37829579 | 0.08 |
ENSRNOT00000084587
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr18_+_60745035 | 0.08 |
ENSRNOT00000045098
|
LOC502176
|
hypothetical protein LOC502176 |
| chr1_-_141504111 | 0.08 |
ENSRNOT00000040164
|
Snrpep2
|
small nuclear ribonucleoprotein polypeptide E pseudogene 2 |
| chrX_-_71169865 | 0.07 |
ENSRNOT00000050415
|
Il2rg
|
interleukin 2 receptor subunit gamma |
| chr3_+_161018511 | 0.07 |
ENSRNOT00000019804
ENSRNOT00000039664 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr1_+_240908483 | 0.07 |
ENSRNOT00000019367
|
Klf9
|
Kruppel-like factor 9 |
| chr5_+_129052043 | 0.07 |
ENSRNOT00000013459
|
LOC108348114
|
tetratricopeptide repeat protein 39A |
| chr3_-_175426395 | 0.07 |
ENSRNOT00000082686
|
Psma7
|
proteasome subunit alpha 7 |
| chr3_+_172195844 | 0.07 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chrX_-_115426083 | 0.07 |
ENSRNOT00000014756
|
AABR07040947.1
|
|
| chrX_-_37353156 | 0.07 |
ENSRNOT00000086120
|
AABR07038021.1
|
|
| chr6_+_55085313 | 0.07 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chr16_+_81665540 | 0.07 |
ENSRNOT00000026546
ENSRNOT00000092644 ENSRNOT00000073240 |
Grtp1
|
growth hormone regulated TBC protein 1 |
| chr12_-_39667849 | 0.06 |
ENSRNOT00000011499
|
Arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr18_+_51523758 | 0.06 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr17_-_90071408 | 0.06 |
ENSRNOT00000071978
|
Pdss1
|
decaprenyl diphosphate synthase subunit 1 |
| chr15_-_23969011 | 0.06 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr1_-_219369587 | 0.06 |
ENSRNOT00000000599
|
Aip
|
aryl-hydrocarbon receptor-interacting protein |
| chr16_+_25773602 | 0.06 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr8_+_132120452 | 0.06 |
ENSRNOT00000055822
|
Tgm4
|
transglutaminase 4 |
| chr8_+_45561390 | 0.06 |
ENSRNOT00000047820
|
AABR07070043.1
|
|
| chr18_-_63825408 | 0.06 |
ENSRNOT00000043709
|
RGD1562758
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr2_+_244521699 | 0.06 |
ENSRNOT00000029382
|
Stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr8_-_112648880 | 0.06 |
ENSRNOT00000015265
|
Ackr4
|
atypical chemokine receptor 4 |
| chr7_-_119158173 | 0.06 |
ENSRNOT00000067483
ENSRNOT00000078528 |
Txn2
|
thioredoxin 2 |
| chr3_-_95007712 | 0.06 |
ENSRNOT00000017409
|
Eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr10_-_49196177 | 0.06 |
ENSRNOT00000084418
|
Zfp286a
|
zinc finger protein 286A |
| chr2_-_189903219 | 0.05 |
ENSRNOT00000017000
|
S100a1
|
S100 calcium binding protein A1 |
| chr19_+_55300395 | 0.05 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chr14_-_115352562 | 0.05 |
ENSRNOT00000050321
|
Erlec1
|
endoplasmic reticulum lectin 1 |
| chr16_+_16949232 | 0.05 |
ENSRNOT00000047499
|
AABR07024795.1
|
|
| chr7_-_29282039 | 0.05 |
ENSRNOT00000067777
|
RGD1561102
|
similar to ribosomal protein S12 |
| chr5_+_156026911 | 0.05 |
ENSRNOT00000046802
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr5_+_115649046 | 0.05 |
ENSRNOT00000041328
|
LOC108351137
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr6_+_128048099 | 0.05 |
ENSRNOT00000084685
ENSRNOT00000087017 |
LOC500712
|
Ab1-233 |
| chr2_-_187863349 | 0.05 |
ENSRNOT00000084455
|
Lmna
|
lamin A/C |
| chr1_+_171592797 | 0.05 |
ENSRNOT00000026607
|
Syt9
|
synaptotagmin 9 |
| chr9_+_17784468 | 0.05 |
ENSRNOT00000026831
|
Slc29a1
|
solute carrier family 29 member 1 |
| chr2_-_166682325 | 0.05 |
ENSRNOT00000091198
ENSRNOT00000012422 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr5_+_57267312 | 0.05 |
ENSRNOT00000011907
|
Chmp5
|
charged multivesicular body protein 5 |
| chr5_-_164747083 | 0.05 |
ENSRNOT00000010433
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr10_-_102289837 | 0.05 |
ENSRNOT00000044922
|
AABR07030729.1
|
|
| chr6_-_78549669 | 0.05 |
ENSRNOT00000009940
|
Foxa1
|
forkhead box A1 |
| chr7_+_14643704 | 0.05 |
ENSRNOT00000044642
|
AABR07055875.1
|
|
| chr9_+_73528681 | 0.04 |
ENSRNOT00000084340
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr9_+_62291809 | 0.04 |
ENSRNOT00000090621
|
Plcl1
|
phospholipase C-like 1 |
| chr5_-_135663284 | 0.04 |
ENSRNOT00000087237
ENSRNOT00000066869 |
Toe1
|
target of EGR1, member 1 (nuclear) |
| chr10_-_95431312 | 0.04 |
ENSRNOT00000085552
|
AABR07030603.2
|
|
| chr7_+_59326518 | 0.04 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr7_+_71023976 | 0.04 |
ENSRNOT00000005679
|
Tac3
|
tachykinin 3 |
| chr1_-_252550394 | 0.04 |
ENSRNOT00000083468
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr1_+_84328114 | 0.04 |
ENSRNOT00000028266
|
Hipk4
|
homeodomain interacting protein kinase 4 |
| chr3_-_105470475 | 0.04 |
ENSRNOT00000011078
|
Gjd2
|
gap junction protein, delta 2 |
| chr1_+_89220083 | 0.04 |
ENSRNOT00000093144
|
Dmkn
|
dermokine |
| chr14_+_2789650 | 0.04 |
ENSRNOT00000030479
|
Fam69a
|
family with sequence similarity 69, member A |
| chr2_+_243656579 | 0.04 |
ENSRNOT00000036993
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr9_-_81565416 | 0.04 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr3_+_122947913 | 0.04 |
ENSRNOT00000028821
|
Vps16
|
VPS16 CORVET/HOPS core subunit |
| chr10_-_85725429 | 0.04 |
ENSRNOT00000005471
|
Rpl23
|
ribosomal protein L23 |
| chr2_+_151314818 | 0.04 |
ENSRNOT00000019305
|
P2ry1
|
purinergic receptor P2Y1 |
| chr9_-_81566096 | 0.04 |
ENSRNOT00000019497
|
Aamp
|
angio-associated, migratory cell protein |
| chr3_-_102826379 | 0.04 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr4_+_159410799 | 0.04 |
ENSRNOT00000079225
|
Akap3
|
A-kinase anchoring protein 3 |
| chr1_-_64030175 | 0.04 |
ENSRNOT00000089950
|
Tsen34l1
|
tRNA splicing endonuclease subunit 34-like 1 |
| chrX_-_123662350 | 0.04 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr19_-_37912027 | 0.04 |
ENSRNOT00000026462
|
Psmb10
|
proteasome subunit beta 10 |
| chr16_-_21089508 | 0.04 |
ENSRNOT00000072565
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chrX_-_45522665 | 0.04 |
ENSRNOT00000030771
|
RGD1562200
|
similar to GS2 gene |
| chr10_-_88533171 | 0.04 |
ENSRNOT00000024321
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chr10_-_89918427 | 0.04 |
ENSRNOT00000084217
|
Dusp3
|
dual specificity phosphatase 3 |
| chr8_+_47674321 | 0.04 |
ENSRNOT00000026170
ENSRNOT00000078776 |
Trim29
|
tripartite motif-containing 29 |
| chr1_+_170147300 | 0.03 |
ENSRNOT00000048075
|
Olr200
|
olfactory receptor 200 |
| chr13_+_90514336 | 0.03 |
ENSRNOT00000088996
ENSRNOT00000085377 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr9_+_98505259 | 0.03 |
ENSRNOT00000051810
|
Erfe
|
erythroferrone |
| chr1_-_196884302 | 0.03 |
ENSRNOT00000089464
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr12_-_23624212 | 0.03 |
ENSRNOT00000064405
ENSRNOT00000001943 |
Rasa4
|
RAS p21 protein activator 4 |
| chr1_-_256813711 | 0.03 |
ENSRNOT00000021055
|
Rbp4
|
retinol binding protein 4 |
| chr3_+_118317761 | 0.03 |
ENSRNOT00000012700
|
Fgf7
|
fibroblast growth factor 7 |
| chr3_+_40038336 | 0.03 |
ENSRNOT00000048804
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr7_-_107616038 | 0.03 |
ENSRNOT00000088752
|
Sla
|
src-like adaptor |
| chr13_-_111765944 | 0.03 |
ENSRNOT00000073041
|
Syt14
|
synaptotagmin 14 |
| chr1_-_201082727 | 0.03 |
ENSRNOT00000079342
|
Ate1
|
arginyltransferase 1 |
| chr11_-_87081950 | 0.03 |
ENSRNOT00000002574
|
Dgcr6
|
DiGeorge syndrome critical region gene 6 |
| chr16_-_64745207 | 0.03 |
ENSRNOT00000032467
|
Tti2
|
TELO2 interacting protein 2 |
| chr12_-_46845079 | 0.03 |
ENSRNOT00000047466
ENSRNOT00000064115 |
Pxn
|
paxillin |
| chr19_+_38669230 | 0.03 |
ENSRNOT00000027273
|
Cdh3
|
cadherin 3 |
| chr3_-_161812243 | 0.03 |
ENSRNOT00000025238
ENSRNOT00000087646 |
Slc35c2
|
solute carrier family 35 member C2 |
| chr5_+_70441123 | 0.03 |
ENSRNOT00000087517
|
Fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr6_-_1319541 | 0.03 |
ENSRNOT00000006527
|
Strn
|
striatin |
| chr8_-_55194692 | 0.03 |
ENSRNOT00000068366
|
RGD1564937
|
similar to RIKEN cDNA 1110032A03 |
| chr4_-_120784807 | 0.03 |
ENSRNOT00000021445
|
Abtb1
|
ankyrin repeat and BTB domain containing 1 |
| chr7_+_29282305 | 0.03 |
ENSRNOT00000007623
|
Arl1
|
ADP-ribosylation factor like GTPase 1 |
| chr10_+_104368247 | 0.03 |
ENSRNOT00000006519
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr10_-_88533829 | 0.03 |
ENSRNOT00000080521
|
Dnajc7
|
DnaJ heat shock protein family (Hsp40) member C7 |
| chrX_+_15295473 | 0.03 |
ENSRNOT00000009295
|
LOC108348065
|
histone deacetylase 6 |
| chr10_+_85646633 | 0.03 |
ENSRNOT00000086608
|
Psmb3
|
proteasome subunit beta 3 |
| chr6_+_55648021 | 0.03 |
ENSRNOT00000064822
ENSRNOT00000091488 |
Ankmy2
|
ankyrin repeat and MYND domain containing 2 |
| chr4_+_129574264 | 0.03 |
ENSRNOT00000010185
|
Arl6ip5
|
ADP-ribosylation factor like GTPase 6 interacting protein 5 |
| chrX_+_159703578 | 0.03 |
ENSRNOT00000001162
|
Cd40lg
|
CD40 ligand |
| chr6_+_128073344 | 0.03 |
ENSRNOT00000014073
|
LOC500712
|
Ab1-233 |
| chr1_-_282251257 | 0.03 |
ENSRNOT00000015186
|
Prdx3
|
peroxiredoxin 3 |
| chr2_-_157759819 | 0.03 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr10_-_18942540 | 0.03 |
ENSRNOT00000007187
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr5_+_148577332 | 0.03 |
ENSRNOT00000016325
|
Snrnp40
|
small nuclear ribonucleoprotein U5 subunit 40 |
| chr5_+_167288717 | 0.03 |
ENSRNOT00000024106
|
Eno1
|
enolase 1 |
| chr2_-_196479770 | 0.02 |
ENSRNOT00000028693
|
Anxa9
|
annexin A9 |
| chr4_+_14039977 | 0.02 |
ENSRNOT00000091249
ENSRNOT00000075878 |
Cd36
|
CD36 molecule |
| chr1_-_57518458 | 0.02 |
ENSRNOT00000002040
|
Pdcd2
|
programmed cell death 2 |
| chr5_+_79367663 | 0.02 |
ENSRNOT00000011508
|
Atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr17_-_21577631 | 0.02 |
ENSRNOT00000066371
|
Tmem14c
|
transmembrane protein 14C |
| chr1_+_142060955 | 0.02 |
ENSRNOT00000017862
|
Vps33b
|
VPS33B, late endosome and lysosome associated |
| chr15_+_7871497 | 0.02 |
ENSRNOT00000046879
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr11_+_75434197 | 0.02 |
ENSRNOT00000032569
|
Mb21d2
|
Mab-21 domain containing 2 |
| chr7_+_14743008 | 0.02 |
ENSRNOT00000055284
|
AABR07055878.1
|
|
| chr1_-_80809769 | 0.02 |
ENSRNOT00000045073
|
Ceacam19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chrX_+_122724129 | 0.02 |
ENSRNOT00000017746
|
LOC100360218
|
interleukin 13 receptor, alpha 1-like |
| chr1_+_42121636 | 0.02 |
ENSRNOT00000025616
|
Myct1
|
myc target 1 |
| chr9_-_70450170 | 0.02 |
ENSRNOT00000017207
|
Mdh1b
|
malate dehydrogenase 1B |
| chr6_-_55647665 | 0.02 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr1_-_153740905 | 0.02 |
ENSRNOT00000023239
|
Prss23
|
protease, serine, 23 |
| chr4_-_165026414 | 0.02 |
ENSRNOT00000071421
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr3_+_93968855 | 0.02 |
ENSRNOT00000014478
|
Fbxo3
|
F-box protein 3 |
| chr8_-_90664554 | 0.02 |
ENSRNOT00000039917
ENSRNOT00000074896 ENSRNOT00000072225 |
Hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr1_-_67065797 | 0.02 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr13_-_99287887 | 0.02 |
ENSRNOT00000004780
|
Ephx1
|
epoxide hydrolase 1 |
| chrX_+_25016401 | 0.02 |
ENSRNOT00000059270
|
Clcn4
|
chloride voltage-gated channel 4 |
| chr10_-_78690857 | 0.02 |
ENSRNOT00000089255
|
LOC303448
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_100832324 | 0.02 |
ENSRNOT00000056364
|
Il4i1
|
interleukin 4 induced 1 |
| chr3_+_79729739 | 0.02 |
ENSRNOT00000084833
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chrX_+_34623405 | 0.02 |
ENSRNOT00000041924
|
Nhs
|
NHS actin remodeling regulator |
| chr10_+_88326080 | 0.02 |
ENSRNOT00000076475
|
Fkbp10
|
FK506 binding protein 10 |
| chr5_-_75319189 | 0.02 |
ENSRNOT00000047200
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr10_-_86355006 | 0.02 |
ENSRNOT00000075076
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr3_-_82777814 | 0.02 |
ENSRNOT00000068585
|
Accsl
|
1-aminocyclopropane-1-carboxylate synthase-like |
| chr8_-_83693472 | 0.02 |
ENSRNOT00000015947
|
Fam83b
|
family with sequence similarity 83, member B |
| chr3_+_132052612 | 0.02 |
ENSRNOT00000030148
|
AABR07053879.1
|
|
| chr8_-_107819050 | 0.02 |
ENSRNOT00000019536
|
Armc8
|
armadillo repeat containing 8 |
| chr4_+_62019970 | 0.02 |
ENSRNOT00000013133
|
LOC100910708
|
aldose reductase-related protein 1-like |
| chr3_-_52998650 | 0.02 |
ENSRNOT00000009988
|
RGD1560831
|
similar to 40S ribosomal protein S3 |
| chr8_-_109576353 | 0.02 |
ENSRNOT00000010320
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr6_+_108167716 | 0.02 |
ENSRNOT00000064426
|
Lin52
|
lin-52 DREAM MuvB core complex component |
| chr7_-_59514939 | 0.02 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr3_+_57770948 | 0.02 |
ENSRNOT00000038429
|
AC107446.2
|
|
| chr8_-_109418872 | 0.02 |
ENSRNOT00000021657
|
Pccb
|
propionyl-CoA carboxylase beta subunit |
| chr9_+_81566074 | 0.02 |
ENSRNOT00000074131
ENSRNOT00000046229 ENSRNOT00000090383 |
Pnkd
|
paroxysmal nonkinesigenic dyskinesia |
| chr2_-_171196395 | 0.02 |
ENSRNOT00000013279
|
Bche
|
butyrylcholinesterase |
| chr1_-_220136470 | 0.02 |
ENSRNOT00000026812
|
Actn3
|
actinin alpha 3 |
| chr4_+_109477920 | 0.02 |
ENSRNOT00000008468
|
Reg3a
|
regenerating islet-derived 3 alpha |
| chrX_+_14358224 | 0.02 |
ENSRNOT00000005042
|
Lancl3
|
LanC like 3 |
| chr13_+_82355886 | 0.02 |
ENSRNOT00000076757
|
Sele
|
selectin E |
| chr20_-_4935372 | 0.02 |
ENSRNOT00000050099
ENSRNOT00000047779 |
RT1-CE3
RT1-CE4
|
RT1 class I, locus CE3 RT1 class I, locus CE4 |
| chr10_+_70242874 | 0.02 |
ENSRNOT00000010924
|
Fndc8
|
fibronectin type III domain containing 8 |
| chr19_-_41029206 | 0.02 |
ENSRNOT00000023383
|
Vac14
|
Vac14, PIKFYVE complex component |
| chr14_-_85398532 | 0.01 |
ENSRNOT00000057361
|
Emid1
|
EMI domain containing 1 |
| chr18_+_30574627 | 0.01 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr16_-_52127591 | 0.01 |
ENSRNOT00000033652
|
Triml2
|
tripartite motif family-like 2 |
| chr4_+_115713004 | 0.01 |
ENSRNOT00000064536
|
Dysf
|
dysferlin |
| chr16_-_49574314 | 0.01 |
ENSRNOT00000017568
ENSRNOT00000085535 ENSRNOT00000017054 |
Pdlim3
|
PDZ and LIM domain 3 |
| chr5_-_148577292 | 0.01 |
ENSRNOT00000017156
|
Zcchc17
|
zinc finger CCHC-type containing 17 |
| chr17_+_57075218 | 0.01 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr13_+_82355471 | 0.01 |
ENSRNOT00000030677
|
Sele
|
selectin E |
| chr6_+_27589657 | 0.01 |
ENSRNOT00000038649
|
Hadha
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit |
| chrX_-_135521598 | 0.01 |
ENSRNOT00000033619
|
Gpr119
|
G protein-coupled receptor 119 |
| chr4_+_115712850 | 0.01 |
ENSRNOT00000085765
|
Dysf
|
dysferlin |
| chr18_+_86307646 | 0.01 |
ENSRNOT00000091778
|
Cd226
|
CD226 molecule |
| chr4_+_119997268 | 0.01 |
ENSRNOT00000073429
|
Rpn1
|
ribophorin I |
| chr18_+_30509393 | 0.01 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr3_+_11476883 | 0.01 |
ENSRNOT00000072241
|
Naif1
|
nuclear apoptosis inducing factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx3-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.1 | 0.4 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 | 0.1 | GO:0052047 | calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0032929 | coumarin metabolic process(GO:0009804) negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.0 | 0.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0042628 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.0 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.0 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.0 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.0 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.0 | GO:0016598 | protein arginylation(GO:0016598) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |