Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
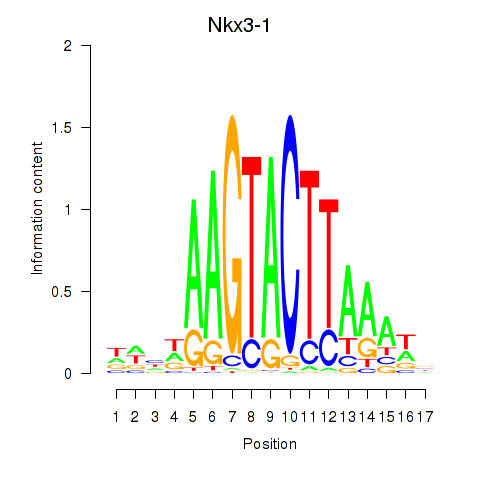
Results for Nkx3-1
Z-value: 0.35
Transcription factors associated with Nkx3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx3-1
|
ENSRNOG00000015477 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx3-1 | rn6_v1_chr15_+_51065316_51065316 | 0.63 | 2.5e-01 | Click! |
Activity profile of Nkx3-1 motif
Sorted Z-values of Nkx3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_80860195 | 0.12 |
ENSRNOT00000038651
|
Trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr3_+_176821652 | 0.10 |
ENSRNOT00000055030
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr13_+_90723092 | 0.09 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr2_+_43271092 | 0.08 |
ENSRNOT00000017571
|
Mier3
|
mesoderm induction early response 1, family member 3 |
| chr10_-_89958805 | 0.08 |
ENSRNOT00000084266
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr12_+_29266602 | 0.08 |
ENSRNOT00000076194
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 |
| chr15_-_55209342 | 0.08 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr18_-_36513317 | 0.07 |
ENSRNOT00000025453
|
Plac8l1
|
PLAC8-like 1 |
| chr6_+_27887797 | 0.07 |
ENSRNOT00000015827
|
Asxl2
|
additional sex combs like 2, transcriptional regulator |
| chr1_+_80954858 | 0.06 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr2_-_49170025 | 0.06 |
ENSRNOT00000060595
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr17_-_32015071 | 0.06 |
ENSRNOT00000070876
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr2_+_55747318 | 0.06 |
ENSRNOT00000050655
|
Dab2
|
DAB2, clathrin adaptor protein |
| chr8_+_19886382 | 0.06 |
ENSRNOT00000034254
|
Zfp317
|
zinc finger protein 317 |
| chr8_-_118696187 | 0.05 |
ENSRNOT00000056141
|
Klhl18
|
kelch-like family member 18 |
| chr8_-_13906355 | 0.05 |
ENSRNOT00000029634
|
Cep295
|
centrosomal protein 295 |
| chr1_-_277939039 | 0.05 |
ENSRNOT00000079579
ENSRNOT00000086726 |
Ablim1
|
actin-binding LIM protein 1 |
| chr2_+_189714754 | 0.05 |
ENSRNOT00000086285
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr8_+_99894762 | 0.05 |
ENSRNOT00000081768
|
Plscr4
|
phospholipid scramblase 4 |
| chr1_+_13595295 | 0.05 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr9_-_65957101 | 0.05 |
ENSRNOT00000014094
|
Mpp4
|
membrane palmitoylated protein 4 |
| chr12_+_37878653 | 0.05 |
ENSRNOT00000091084
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr17_-_89923423 | 0.05 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr1_+_263448633 | 0.05 |
ENSRNOT00000064510
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr16_-_21473808 | 0.05 |
ENSRNOT00000066776
|
Zfp964
|
zinc finger protein 964 |
| chrX_+_15114892 | 0.05 |
ENSRNOT00000078168
|
Wdr13
|
WD repeat domain 13 |
| chr19_+_9668186 | 0.05 |
ENSRNOT00000016563
|
Cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr5_-_166695134 | 0.05 |
ENSRNOT00000047763
|
Tmem201
|
transmembrane protein 201 |
| chr17_-_69862110 | 0.05 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chrX_+_159081445 | 0.04 |
ENSRNOT00000056688
|
AABR07042542.1
|
|
| chr8_-_36388224 | 0.04 |
ENSRNOT00000038312
|
Tirap
|
TIR domain containing adaptor protein |
| chr15_-_88670349 | 0.04 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr14_+_108415068 | 0.04 |
ENSRNOT00000083763
|
Pus10
|
pseudouridylate synthase 10 |
| chr3_+_112371677 | 0.04 |
ENSRNOT00000013316
|
LOC100912076
|
HAUS augmin-like complex subunit 2-like |
| chr8_-_93390305 | 0.04 |
ENSRNOT00000056930
|
Ibtk
|
inhibitor of Bruton tyrosine kinase |
| chr16_+_20039969 | 0.04 |
ENSRNOT00000024866
|
Colgalt1
|
collagen beta(1-O)galactosyltransferase 1 |
| chr6_+_126874193 | 0.04 |
ENSRNOT00000011775
|
Unc79
|
unc-79 homolog (C. elegans) |
| chr7_-_134482607 | 0.04 |
ENSRNOT00000081104
|
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr6_+_26390689 | 0.04 |
ENSRNOT00000079762
ENSRNOT00000091528 |
Ift172
|
intraflagellar transport 172 |
| chr18_+_30904498 | 0.04 |
ENSRNOT00000026969
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr18_-_35817117 | 0.04 |
ENSRNOT00000015172
ENSRNOT00000085714 |
Mcc
|
mutated in colorectal cancers |
| chr15_+_47455690 | 0.04 |
ENSRNOT00000075276
|
LOC100910401
|
serine protease 55-like |
| chr6_+_2216623 | 0.04 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr16_+_53998560 | 0.04 |
ENSRNOT00000077188
ENSRNOT00000013463 |
Asah1
|
N-acylsphingosine amidohydrolase 1 |
| chr3_+_112385601 | 0.04 |
ENSRNOT00000056210
|
LOC691918
|
similar to Centrosomal protein of 27 kDa (Cep27 protein) |
| chr13_+_98311827 | 0.04 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_+_172155496 | 0.04 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
| chr5_+_135413275 | 0.04 |
ENSRNOT00000012263
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chrX_-_40086870 | 0.04 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr5_+_139783951 | 0.04 |
ENSRNOT00000081333
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr13_+_71110064 | 0.04 |
ENSRNOT00000088329
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr13_-_26769374 | 0.04 |
ENSRNOT00000003768
|
Bcl2
|
BCL2, apoptosis regulator |
| chr3_-_112371664 | 0.04 |
ENSRNOT00000079606
ENSRNOT00000012636 |
LOC100910478
|
leucine-rich repeat-containing protein 57-like |
| chr3_+_151688454 | 0.04 |
ENSRNOT00000026856
|
Romo1
|
reactive oxygen species modulator 1 |
| chr3_+_80468154 | 0.03 |
ENSRNOT00000021837
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr2_+_197720259 | 0.03 |
ENSRNOT00000070919
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr6_+_86872903 | 0.03 |
ENSRNOT00000086894
|
Fancm
|
Fanconi anemia, complementation group M |
| chr6_+_99625306 | 0.03 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr1_+_156552328 | 0.03 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr7_+_2645196 | 0.03 |
ENSRNOT00000083899
|
Timeless
|
timeless circadian clock |
| chr16_+_54332660 | 0.03 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr17_-_42226377 | 0.03 |
ENSRNOT00000024536
|
RGD1307443
|
similar to mKIAA0319 protein |
| chr9_+_71230108 | 0.03 |
ENSRNOT00000018326
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr15_+_27933211 | 0.03 |
ENSRNOT00000013619
|
Rnase10
|
ribonuclease A family member 10 |
| chr3_-_112385528 | 0.03 |
ENSRNOT00000049756
|
Lrrc57
|
leucine rich repeat containing 57 |
| chr2_+_86914989 | 0.03 |
ENSRNOT00000085164
ENSRNOT00000081966 |
LOC100360380
|
zinc finger protein 457-like |
| chr2_-_54963448 | 0.03 |
ENSRNOT00000017886
|
Ptger4
|
prostaglandin E receptor 4 |
| chr9_-_81772851 | 0.03 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr7_-_139542461 | 0.03 |
ENSRNOT00000089425
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr14_+_106153575 | 0.03 |
ENSRNOT00000010264
|
Vps54
|
VPS54 GARP complex subunit |
| chr4_+_160020472 | 0.03 |
ENSRNOT00000078802
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr1_-_67134827 | 0.03 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr10_-_36419926 | 0.03 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr16_-_9658484 | 0.03 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr7_+_54031316 | 0.03 |
ENSRNOT00000039096
|
Bbs10
|
Bardet-Biedl syndrome 10 |
| chr3_-_1868015 | 0.03 |
ENSRNOT00000048945
|
Cacna1b
|
calcium voltage-gated channel subunit alpha1 B |
| chr14_+_77322012 | 0.03 |
ENSRNOT00000088600
ENSRNOT00000041639 |
Zbtb49
|
zinc finger and BTB domain containing 49 |
| chr5_-_159293673 | 0.03 |
ENSRNOT00000009081
|
Padi4
|
peptidyl arginine deiminase 4 |
| chr5_+_6373583 | 0.03 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr4_-_176596591 | 0.03 |
ENSRNOT00000086365
|
Recql
|
RecQ like helicase |
| chr13_+_105684420 | 0.03 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr19_+_46455761 | 0.03 |
ENSRNOT00000034012
ENSRNOT00000093392 |
Nudt7
|
nudix hydrolase 7 |
| chr13_+_89524329 | 0.02 |
ENSRNOT00000004279
|
Mpz
|
myelin protein zero |
| chr2_+_143895982 | 0.02 |
ENSRNOT00000080026
|
Supt20h
|
SPT20 homolog, SAGA complex component |
| chr3_-_64554953 | 0.02 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
| chr11_+_87224421 | 0.02 |
ENSRNOT00000000308
|
Dgcr14
|
DiGeorge syndrome critical region gene 14 |
| chr2_-_231648122 | 0.02 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr10_+_13145099 | 0.02 |
ENSRNOT00000091287
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr7_+_40217991 | 0.02 |
ENSRNOT00000085684
|
Cep290
|
centrosomal protein 290 |
| chr3_-_71798531 | 0.02 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr13_-_82295123 | 0.02 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chrX_+_9436707 | 0.02 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr2_-_39007976 | 0.02 |
ENSRNOT00000064292
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr14_+_81858737 | 0.02 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr8_-_78397123 | 0.02 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr5_+_104394119 | 0.02 |
ENSRNOT00000093557
ENSRNOT00000093462 |
Adamtsl1
|
ADAMTS-like 1 |
| chr6_-_42302254 | 0.02 |
ENSRNOT00000007071
|
Pqlc3
|
PQ loop repeat containing 3 |
| chr17_-_58301260 | 0.02 |
ENSRNOT00000083331
|
Adarb2
|
adenosine deaminase, RNA-specific, B2 |
| chr10_-_70181708 | 0.02 |
ENSRNOT00000076598
|
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr14_+_48726045 | 0.02 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr1_-_73399579 | 0.02 |
ENSRNOT00000077186
|
Lilrb4
|
leukocyte immunoglobulin like receptor B4 |
| chr8_+_116826680 | 0.02 |
ENSRNOT00000026854
|
Gmppb
|
GDP-mannose pyrophosphorylase B |
| chr3_-_111266542 | 0.02 |
ENSRNOT00000031672
|
Ino80
|
INO80 complex subunit |
| chr1_+_253221812 | 0.02 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr11_+_42936028 | 0.02 |
ENSRNOT00000074653
|
AABR07033887.1
|
|
| chr7_+_74350479 | 0.02 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr10_-_70681672 | 0.02 |
ENSRNOT00000013836
|
Mmp28
|
matrix metallopeptidase 28 |
| chr14_+_66631690 | 0.02 |
ENSRNOT00000079658
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr18_+_30820321 | 0.02 |
ENSRNOT00000060472
|
Pcdhga3
|
protocadherin gamma subfamily A, 3 |
| chr8_+_111210811 | 0.02 |
ENSRNOT00000011347
|
Amotl2
|
angiomotin like 2 |
| chr6_+_110410141 | 0.02 |
ENSRNOT00000013867
|
Esrrb
|
estrogen-related receptor beta |
| chr8_-_63750531 | 0.02 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr10_-_65502936 | 0.02 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr4_-_16669368 | 0.02 |
ENSRNOT00000007608
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr4_+_142780987 | 0.02 |
ENSRNOT00000056570
|
Grm7
|
glutamate metabotropic receptor 7 |
| chr2_-_184951950 | 0.02 |
ENSRNOT00000093486
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr5_-_134776247 | 0.02 |
ENSRNOT00000057055
|
Dmbx1
|
diencephalon/mesencephalon homeobox 1 |
| chr2_-_203413124 | 0.02 |
ENSRNOT00000077898
ENSRNOT00000056140 |
Cd101
|
CD101 molecule |
| chr6_-_2316717 | 0.02 |
ENSRNOT00000082017
ENSRNOT00000066363 |
Cyp1b1
|
cytochrome P450, family 1, subfamily b, polypeptide 1 |
| chr1_+_84758233 | 0.02 |
ENSRNOT00000072734
|
AABR07002775.1
|
|
| chr1_+_30863217 | 0.02 |
ENSRNOT00000015421
|
Rnf146
|
ring finger protein 146 |
| chr6_+_102477036 | 0.02 |
ENSRNOT00000087132
|
Rad51b
|
RAD51 paralog B |
| chrX_+_113510621 | 0.02 |
ENSRNOT00000025912
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chrX_-_158261717 | 0.02 |
ENSRNOT00000086804
|
RGD1561558
|
similar to RIKEN cDNA 1700001F22 |
| chr14_+_3204390 | 0.02 |
ENSRNOT00000088078
|
Glmn
|
glomulin, FKBP associated protein |
| chr13_-_95348913 | 0.02 |
ENSRNOT00000057879
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr2_-_165591110 | 0.02 |
ENSRNOT00000091140
|
Ift80
|
intraflagellar transport 80 |
| chr8_-_39201588 | 0.02 |
ENSRNOT00000011226
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr8_+_117600749 | 0.02 |
ENSRNOT00000047482
|
Nckipsd
|
NCK interacting protein with SH3 domain |
| chr1_+_255564409 | 0.02 |
ENSRNOT00000081786
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr4_+_22898527 | 0.02 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr19_+_38162515 | 0.02 |
ENSRNOT00000027117
|
Slc7a6
|
solute carrier family 7 member 6 |
| chr3_-_21609853 | 0.02 |
ENSRNOT00000012160
|
Pdcl
|
phosducin-like |
| chr13_-_15395617 | 0.02 |
ENSRNOT00000046060
|
LOC304725
|
similar to contactin associated protein-like 5 isoform 1 |
| chr9_-_23352668 | 0.02 |
ENSRNOT00000075279
|
Mut
|
methylmalonyl CoA mutase |
| chrM_+_9870 | 0.02 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr4_+_117743710 | 0.02 |
ENSRNOT00000021491
|
Add2
|
adducin 2 |
| chrX_+_157759624 | 0.02 |
ENSRNOT00000003192
|
LOC686087
|
similar to motile sperm domain containing 1 |
| chr12_+_12859661 | 0.02 |
ENSRNOT00000001404
|
Usp42
|
ubiquitin specific peptidase 42 |
| chr8_+_111209908 | 0.01 |
ENSRNOT00000090221
|
Amotl2
|
angiomotin like 2 |
| chr18_+_3163214 | 0.01 |
ENSRNOT00000017291
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr12_+_2140203 | 0.01 |
ENSRNOT00000084906
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr15_-_33285779 | 0.01 |
ENSRNOT00000036150
|
Cdh24
|
cadherin 24 |
| chr8_-_49269411 | 0.01 |
ENSRNOT00000021321
|
Ube4a
|
ubiquitination factor E4A |
| chr7_-_68523364 | 0.01 |
ENSRNOT00000077670
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr1_-_126211439 | 0.01 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr4_+_70572942 | 0.01 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr1_-_67065797 | 0.01 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr2_-_220535751 | 0.01 |
ENSRNOT00000089082
|
Palmd
|
palmdelphin |
| chr6_-_27460038 | 0.01 |
ENSRNOT00000036815
|
Drc1
|
dynein regulatory complex subunit 1 |
| chr2_+_53109684 | 0.01 |
ENSRNOT00000086590
|
Selenop
|
selenoprotein P |
| chrX_+_68627313 | 0.01 |
ENSRNOT00000076699
ENSRNOT00000076795 ENSRNOT00000008705 |
Yipf6
|
Yip1 domain family, member 6 |
| chr1_+_85162452 | 0.01 |
ENSRNOT00000093347
|
Pak4
|
p21 (RAC1) activated kinase 4 |
| chr10_-_31419235 | 0.01 |
ENSRNOT00000059496
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr17_+_22620721 | 0.01 |
ENSRNOT00000019478
|
Adtrp
|
androgen-dependent TFPI-regulating protein |
| chr1_+_41590494 | 0.01 |
ENSRNOT00000089984
|
Esr1
|
estrogen receptor 1 |
| chr2_+_127625683 | 0.01 |
ENSRNOT00000015474
|
Hspa4l
|
heat shock protein 4-like |
| chr16_-_54137660 | 0.01 |
ENSRNOT00000085435
|
Pcm1
|
pericentriolar material 1 |
| chr10_+_70520206 | 0.01 |
ENSRNOT00000088198
ENSRNOT00000090446 ENSRNOT00000085799 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr8_-_27852996 | 0.01 |
ENSRNOT00000037790
|
Glb1l2
|
galactosidase, beta 1-like 2 |
| chr19_+_37990374 | 0.01 |
ENSRNOT00000026827
|
Dus2
|
dihydrouridine synthase 2 |
| chr1_-_255976425 | 0.01 |
ENSRNOT00000085303
|
Ide
|
insulin degrading enzyme |
| chr2_-_211715311 | 0.01 |
ENSRNOT00000027738
|
Prpf38b
|
pre-mRNA processing factor 38B |
| chr12_+_10577068 | 0.01 |
ENSRNOT00000001282
|
Rnf6
|
ring finger protein 6 |
| chr10_+_90985653 | 0.01 |
ENSRNOT00000093364
|
Ccdc103
|
coiled-coil domain containing 103 |
| chr3_-_105470475 | 0.01 |
ENSRNOT00000011078
|
Gjd2
|
gap junction protein, delta 2 |
| chr11_+_20474483 | 0.01 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chrX_+_29504349 | 0.01 |
ENSRNOT00000005981
|
Tceanc
|
transcription elongation factor A N-terminal and central domain containing |
| chrX_-_64726210 | 0.01 |
ENSRNOT00000076012
ENSRNOT00000086265 |
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr8_+_103342929 | 0.01 |
ENSRNOT00000045455
|
LOC501038
|
Ab2-060 |
| chr13_-_47623849 | 0.01 |
ENSRNOT00000006106
|
Il24
|
interleukin 24 |
| chr4_-_140220487 | 0.01 |
ENSRNOT00000009008
|
Sumf1
|
sulfatase modifying factor 1 |
| chr14_-_45859908 | 0.01 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr18_+_15738809 | 0.01 |
ENSRNOT00000075444
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr10_+_67810810 | 0.01 |
ENSRNOT00000079156
|
Psmd11
|
proteasome 26S subunit, non-ATPase 11 |
| chr20_-_27117663 | 0.01 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chr5_-_150949931 | 0.01 |
ENSRNOT00000067905
|
Smpdl3b
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_-_27094364 | 0.01 |
ENSRNOT00000050902
|
Rwdd2b
|
RWD domain containing 2B |
| chr20_+_3246739 | 0.01 |
ENSRNOT00000061299
|
RT1-T24-2
|
RT1 class I, locus T24, gene 2 |
| chr16_-_54137485 | 0.01 |
ENSRNOT00000014202
|
Pcm1
|
pericentriolar material 1 |
| chr5_-_115184219 | 0.01 |
ENSRNOT00000045686
|
LOC100912642
|
cytochrome P450 2J3-like |
| chr1_-_198354466 | 0.01 |
ENSRNOT00000027094
|
Tmem219
|
transmembrane protein 219 |
| chr13_+_73708815 | 0.01 |
ENSRNOT00000071482
ENSRNOT00000077874 ENSRNOT00000050734 |
Tor1aip2
Tor1aip2
|
torsin 1A interacting protein 2 torsin 1A interacting protein 2 |
| chr18_-_81682206 | 0.01 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr1_-_277867630 | 0.01 |
ENSRNOT00000054679
ENSRNOT00000091038 ENSRNOT00000079869 |
Ablim1
|
actin-binding LIM protein 1 |
| chr19_+_43392085 | 0.01 |
ENSRNOT00000023854
|
Sf3b3
|
splicing factor 3b, subunit 3 |
| chr8_-_132911193 | 0.01 |
ENSRNOT00000087799
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr1_-_66639205 | 0.01 |
ENSRNOT00000082190
|
LOC100909481
|
eukaryotic translation initiation factor 3 subunit E-like |
| chr9_-_15700235 | 0.01 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr1_+_7252349 | 0.01 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr9_-_27761365 | 0.01 |
ENSRNOT00000018552
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr10_+_51478378 | 0.01 |
ENSRNOT00000004722
|
Elac2
|
elaC ribonuclease Z 2 |
| chr18_+_56887722 | 0.01 |
ENSRNOT00000078783
|
Csnk1a1
|
casein kinase 1, alpha 1 |
| chr10_+_104377748 | 0.01 |
ENSRNOT00000080714
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr10_+_17579284 | 0.01 |
ENSRNOT00000079698
|
Fbxw11
|
F-box and WD repeat domain containing 11 |
| chr6_-_92398525 | 0.01 |
ENSRNOT00000007314
|
Sav1
|
salvador family WW domain containing protein 1 |
| chr2_-_60461458 | 0.01 |
ENSRNOT00000024285
|
Brix1
|
BRX1, biogenesis of ribosomes |
| chr4_+_342302 | 0.01 |
ENSRNOT00000009233
|
Insig1
|
insulin induced gene 1 |
| chr10_+_72147816 | 0.01 |
ENSRNOT00000090085
|
LOC102553386
|
SUMO-conjugating enzyme UBC9-like |
| chr8_+_13305152 | 0.01 |
ENSRNOT00000012940
|
Mre11a
|
MRE11 homolog A, double strand break repair nuclease |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:1903943 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.0 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:0046671 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.0 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.0 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |