Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
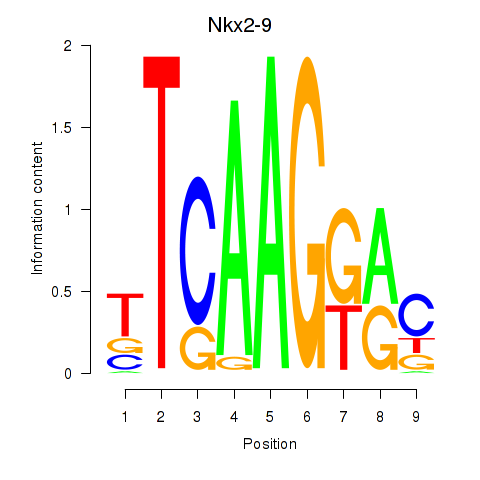
Results for Nkx2-9
Z-value: 0.46
Transcription factors associated with Nkx2-9
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Nkx2-9 motif
Sorted Z-values of Nkx2-9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_28654733 | 0.81 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr1_+_33910912 | 0.24 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr1_-_7801438 | 0.17 |
ENSRNOT00000022273
|
AABR07000276.1
|
|
| chr5_+_50381244 | 0.17 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr1_+_279798187 | 0.15 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr5_-_127273656 | 0.14 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr7_-_119441487 | 0.13 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr8_-_61917125 | 0.11 |
ENSRNOT00000085049
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr10_-_87407634 | 0.11 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr3_+_161018511 | 0.11 |
ENSRNOT00000019804
ENSRNOT00000039664 |
Wfdc2
|
WAP four-disulfide core domain 2 |
| chr4_-_30338679 | 0.10 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr6_-_145777767 | 0.10 |
ENSRNOT00000046799
|
Rpl32
|
ribosomal protein L32 |
| chr5_-_38923095 | 0.09 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr11_+_81358592 | 0.09 |
ENSRNOT00000002487
|
Rfc4
|
replication factor C subunit 4 |
| chrM_+_7758 | 0.08 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr14_-_44375804 | 0.08 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr9_-_29337922 | 0.08 |
ENSRNOT00000018896
|
RGD1309049
|
similar to RIKEN cDNA 4933415F23 |
| chr20_-_27117663 | 0.07 |
ENSRNOT00000000434
|
Pbld1
|
phenazine biosynthesis-like protein domain containing 1 |
| chrX_+_77065397 | 0.07 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr9_+_69953440 | 0.07 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_256786124 | 0.07 |
ENSRNOT00000034563
|
Ffar4
|
free fatty acid receptor 4 |
| chr8_-_43480777 | 0.07 |
ENSRNOT00000060074
|
LOC688473
|
similar to 40S ribosomal protein S2 |
| chr3_-_2798470 | 0.07 |
ENSRNOT00000022495
|
Phpt1
|
phosphohistidine phosphatase 1 |
| chr12_+_22274828 | 0.07 |
ENSRNOT00000001914
|
Epo
|
erythropoietin |
| chr1_-_84812486 | 0.06 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr3_-_60813869 | 0.06 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr12_-_30566032 | 0.06 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr11_+_60072727 | 0.06 |
ENSRNOT00000090230
|
Tagln3
|
transgelin 3 |
| chr11_+_57505005 | 0.06 |
ENSRNOT00000002942
|
LOC103693564
|
transgelin-3 |
| chr7_+_98813040 | 0.06 |
ENSRNOT00000012616
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr6_-_93562314 | 0.06 |
ENSRNOT00000010871
ENSRNOT00000088790 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr12_+_44046217 | 0.05 |
ENSRNOT00000080853
ENSRNOT00000079510 |
Fbxw8
|
F-box and WD repeat domain containing 8 |
| chr1_-_233140237 | 0.05 |
ENSRNOT00000083372
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr13_+_42220251 | 0.05 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr14_+_46649971 | 0.04 |
ENSRNOT00000085875
|
AABR07015078.1
|
|
| chr4_-_176720012 | 0.04 |
ENSRNOT00000017965
|
Ldhb
|
lactate dehydrogenase B |
| chr8_+_89369417 | 0.04 |
ENSRNOT00000037530
|
Mei4
|
meiotic double-stranded break formation protein 4 |
| chr4_+_145413230 | 0.04 |
ENSRNOT00000056508
|
Il17re
|
interleukin 17 receptor E |
| chr13_+_51972974 | 0.04 |
ENSRNOT00000008164
|
Arl8a
|
ADP-ribosylation factor like GTPase 8A |
| chr7_-_55604403 | 0.04 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr15_-_29369504 | 0.04 |
ENSRNOT00000060297
|
AABR07017624.1
|
|
| chr8_+_48406260 | 0.04 |
ENSRNOT00000009975
|
Usp2
|
ubiquitin specific peptidase 2 |
| chrX_-_15467875 | 0.04 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr8_+_62283336 | 0.04 |
ENSRNOT00000025410
|
Rpp25
|
ribonuclease P/MRP 25 subunit |
| chr8_-_115626380 | 0.04 |
ENSRNOT00000019107
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr9_-_17254949 | 0.04 |
ENSRNOT00000026499
|
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr2_+_251983339 | 0.03 |
ENSRNOT00000020230
|
Mcoln3
|
mucolipin 3 |
| chr4_-_145390447 | 0.03 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr6_+_58467254 | 0.03 |
ENSRNOT00000065396
|
Etv1
|
ets variant 1 |
| chrX_+_128493614 | 0.03 |
ENSRNOT00000044240
|
Stag2
|
stromal antigen 2 |
| chrM_+_7919 | 0.03 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr4_-_131694755 | 0.03 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr9_+_66058047 | 0.03 |
ENSRNOT00000071819
|
Cdk15
|
cyclin-dependent kinase 15 |
| chr10_-_31493419 | 0.03 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chr14_+_108826831 | 0.03 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr2_+_205525204 | 0.03 |
ENSRNOT00000091210
|
Csde1
|
cold shock domain containing E1 |
| chr15_-_28696122 | 0.03 |
ENSRNOT00000060467
|
Rab2b
|
RAB2B, member RAS oncogene family |
| chr7_+_35773928 | 0.03 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr6_-_138662365 | 0.03 |
ENSRNOT00000066209
ENSRNOT00000084892 |
Ighm
|
immunoglobulin heavy constant mu |
| chr2_-_23260965 | 0.03 |
ENSRNOT00000046772
|
LOC103691423
|
60S ribosomal protein L36 pseudogene |
| chrX_-_16526028 | 0.03 |
ENSRNOT00000093647
|
Dgkk
|
diacylglycerol kinase kappa |
| chr1_-_90091287 | 0.02 |
ENSRNOT00000032613
|
Gpi
|
glucose-6-phosphate isomerase |
| chr2_-_241472694 | 0.02 |
ENSRNOT00000051498
|
Bank1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr3_-_113342675 | 0.02 |
ENSRNOT00000020130
|
Strc
|
stereocilin |
| chr20_+_41184287 | 0.02 |
ENSRNOT00000081117
|
Col10a1
|
collagen type X alpha 1 chain |
| chrX_-_152367861 | 0.02 |
ENSRNOT00000089414
|
LOC103690371
|
melanoma-associated antigen 10-like |
| chr1_-_174047572 | 0.02 |
ENSRNOT00000019597
|
Stk33
|
serine/threonine kinase 33 |
| chr12_+_48101673 | 0.02 |
ENSRNOT00000065060
|
Foxn4
|
forkhead box N4 |
| chr17_+_54280851 | 0.02 |
ENSRNOT00000024022
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr10_+_15241590 | 0.02 |
ENSRNOT00000037372
ENSRNOT00000037381 |
Mettl26
|
methyltransferase like 26 |
| chr7_+_141355994 | 0.02 |
ENSRNOT00000081195
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chrX_-_64908682 | 0.02 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr19_-_14945302 | 0.02 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr5_-_107388182 | 0.01 |
ENSRNOT00000058358
|
LOC100912356
|
interferon alpha-1-like |
| chr9_+_81644355 | 0.01 |
ENSRNOT00000071700
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr15_+_28696378 | 0.01 |
ENSRNOT00000017476
|
Tox4
|
TOX high mobility group box family member 4 |
| chr20_-_4132604 | 0.01 |
ENSRNOT00000077630
ENSRNOT00000048332 |
RT1-Da
|
RT1 class II, locus Da |
| chr20_+_3364814 | 0.01 |
ENSRNOT00000001077
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chrX_-_81817101 | 0.01 |
ENSRNOT00000003730
|
AABR07039648.1
|
|
| chr5_-_153184940 | 0.01 |
ENSRNOT00000079536
|
Tmem57
|
transmembrane protein 57 |
| chr15_+_58170967 | 0.01 |
ENSRNOT00000001366
|
Nufip1
|
NUFIP1, FMR1 interacting protein 1 |
| chr8_-_39266959 | 0.01 |
ENSRNOT00000046590
|
Ei24
|
EI24, autophagy associated transmembrane protein |
| chr18_-_52017734 | 0.01 |
ENSRNOT00000081020
|
March3
|
membrane associated ring-CH-type finger 3 |
| chr1_-_90961717 | 0.01 |
ENSRNOT00000071180
|
LOC108348106
|
alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 |
| chr5_-_73494030 | 0.01 |
ENSRNOT00000022291
|
Actl7b
|
actin-like 7b |
| chr1_-_197778490 | 0.01 |
ENSRNOT00000024185
|
Spns1
|
SPNS sphingolipid transporter 1 |
| chr7_+_142106465 | 0.01 |
ENSRNOT00000042786
ENSRNOT00000088681 |
Letmd1
|
LETM1 domain containing 1 |
| chr17_-_52569036 | 0.01 |
ENSRNOT00000019396
|
Gli3
|
GLI family zinc finger 3 |
| chr17_+_47721977 | 0.01 |
ENSRNOT00000080800
|
LOC100910792
|
amphiphysin-like |
| chr18_+_30010918 | 0.01 |
ENSRNOT00000084132
|
Pcdha4
|
protocadherin alpha 4 |
| chr19_-_41010122 | 0.00 |
ENSRNOT00000088770
|
Vac14
|
Vac14, PIKFYVE complex component |
| chr11_-_67037115 | 0.00 |
ENSRNOT00000003137
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chrX_-_23649466 | 0.00 |
ENSRNOT00000059431
|
Shroom2
|
shroom family member 2 |
| chr8_+_50525091 | 0.00 |
ENSRNOT00000074357
|
Apoa1
|
apolipoprotein A1 |
| chr9_+_81427730 | 0.00 |
ENSRNOT00000019109
ENSRNOT00000081711 |
Cxcr2
|
C-X-C motif chemokine receptor 2 |
| chrX_+_71155894 | 0.00 |
ENSRNOT00000076479
|
Foxo4
|
forkhead box O4 |
| chr8_+_85951191 | 0.00 |
ENSRNOT00000037665
|
Cd109
|
CD109 molecule |
| chr1_+_46779860 | 0.00 |
ENSRNOT00000075268
|
Snx9
|
sorting nexin 9 |
| chr12_-_11144754 | 0.00 |
ENSRNOT00000084637
|
Zfp655
|
zinc finger protein 655 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.2 | GO:0072268 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0046884 | thyroid hormone generation(GO:0006590) follicle-stimulating hormone secretion(GO:0046884) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |