Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Nkx2-5
Z-value: 0.30
Transcription factors associated with Nkx2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-5
|
ENSRNOG00000020747 | NK2 homeobox 5 |
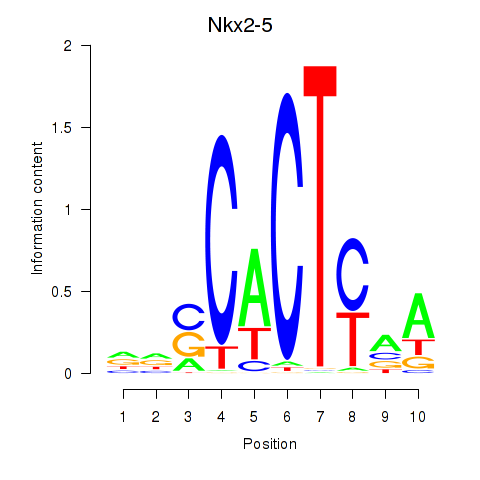
Activity profile of Nkx2-5 motif
Sorted Z-values of Nkx2-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_106153045 | 0.10 |
ENSRNOT00000092065
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr5_-_83648044 | 0.09 |
ENSRNOT00000046725
|
RGD1561195
|
similar to ribosomal protein L31 |
| chr12_-_19254527 | 0.08 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chr11_+_45888221 | 0.06 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr13_-_107886476 | 0.06 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr9_-_60923706 | 0.05 |
ENSRNOT00000047517
|
AABR07067749.1
|
|
| chr13_-_82005741 | 0.05 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr15_-_14510828 | 0.05 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr5_+_164808323 | 0.05 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr1_+_87248489 | 0.04 |
ENSRNOT00000028091
|
Dpf1
|
double PHD fingers 1 |
| chr11_-_61530567 | 0.04 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr20_-_4390436 | 0.04 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr6_+_57516713 | 0.04 |
ENSRNOT00000063874
|
Dgkb
|
diacylglycerol kinase, beta |
| chr8_-_43480777 | 0.04 |
ENSRNOT00000060074
|
LOC688473
|
similar to 40S ribosomal protein S2 |
| chr10_-_43512287 | 0.03 |
ENSRNOT00000036481
|
Faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr2_-_198458041 | 0.03 |
ENSRNOT00000082450
|
Fcgr1a
|
Fc fragment of IgG receptor Ia |
| chr14_-_114047527 | 0.03 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr10_+_49472460 | 0.03 |
ENSRNOT00000038276
|
Tekt3
|
tektin 3 |
| chr16_-_64806050 | 0.03 |
ENSRNOT00000015725
|
Dusp26
|
dual specificity phosphatase 26 |
| chr12_-_13462038 | 0.03 |
ENSRNOT00000043110
|
LOC100360439
|
ribosomal protein L36-like |
| chr7_-_35550247 | 0.03 |
ENSRNOT00000049211
|
Rpl31l3
|
ribosomal protein L31-like 3 |
| chr10_-_65820538 | 0.03 |
ENSRNOT00000011808
|
Tmem97
|
transmembrane protein 97 |
| chr7_-_120558805 | 0.03 |
ENSRNOT00000087344
|
Pla2g6
|
phospholipase A2 group VI |
| chr7_-_120559157 | 0.03 |
ENSRNOT00000090565
ENSRNOT00000016827 ENSRNOT00000017108 |
Pla2g6
|
phospholipase A2 group VI |
| chr10_-_83655182 | 0.03 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr8_-_53758774 | 0.03 |
ENSRNOT00000068040
|
Ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr2_-_185444897 | 0.03 |
ENSRNOT00000016329
|
Rps3a
|
ribosomal protein S3a |
| chr6_+_115170865 | 0.03 |
ENSRNOT00000005671
|
Tshr
|
thyroid stimulating hormone receptor |
| chr18_+_56071478 | 0.03 |
ENSRNOT00000025344
ENSRNOT00000025354 |
Cd74
|
CD74 molecule |
| chr7_-_119797098 | 0.02 |
ENSRNOT00000009994
|
Rac2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr16_+_49185560 | 0.02 |
ENSRNOT00000014360
|
Helt
|
helt bHLH transcription factor |
| chr14_-_85088523 | 0.02 |
ENSRNOT00000010690
|
Nf2
|
neurofibromin 2 |
| chr9_+_44567206 | 0.02 |
ENSRNOT00000088544
|
NEWGENE_1308196
|
mitochondrial ribosomal protein L30 |
| chr2_-_23260965 | 0.02 |
ENSRNOT00000046772
|
LOC103691423
|
60S ribosomal protein L36 pseudogene |
| chr2_+_252452269 | 0.02 |
ENSRNOT00000021970
|
Uox
|
urate oxidase |
| chr5_-_32956159 | 0.02 |
ENSRNOT00000078264
|
Cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr9_-_10182676 | 0.02 |
ENSRNOT00000074894
|
LOC316124
|
similar to gonadotropin-regulated long chain acyl-CoA synthetase |
| chr5_-_155709215 | 0.02 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chr16_-_18819833 | 0.02 |
ENSRNOT00000072664
|
F2rl3
|
F2R like thrombin/trypsin receptor 3 |
| chr17_-_36438752 | 0.02 |
ENSRNOT00000048997
|
RGD1561310
|
similar to ribosomal protein L37 |
| chr12_-_19159901 | 0.01 |
ENSRNOT00000045401
|
Rpl31l4
|
ribosomal protein L31-like 4 |
| chr5_-_78393143 | 0.01 |
ENSRNOT00000043610
|
LOC100910017
|
60S ribosomal protein L31-like |
| chr16_+_50049828 | 0.01 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr7_+_107467260 | 0.01 |
ENSRNOT00000009240
ENSRNOT00000077382 |
Tg
|
thyroglobulin |
| chr9_+_4817854 | 0.01 |
ENSRNOT00000040879
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr2_-_252451999 | 0.01 |
ENSRNOT00000021861
|
Dnase2b
|
deoxyribonuclease 2 beta |
| chr1_+_61132818 | 0.01 |
ENSRNOT00000041434
|
Vom1r20
|
vomeronasal 1 receptor 20 |
| chr1_-_89258935 | 0.01 |
ENSRNOT00000003180
|
LOC100361079
|
ribosomal protein L36-like |
| chr5_+_58937615 | 0.01 |
ENSRNOT00000080909
|
Tesk1
|
testis-specific kinase 1 |
| chr6_+_58388333 | 0.01 |
ENSRNOT00000036337
|
LOC680646
|
similar to 40S ribosomal protein S2 |
| chr10_-_53595827 | 0.01 |
ENSRNOT00000004560
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr15_+_23777856 | 0.01 |
ENSRNOT00000060847
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chrX_+_37329779 | 0.01 |
ENSRNOT00000038352
ENSRNOT00000088802 |
Pdha1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr20_-_4391402 | 0.01 |
ENSRNOT00000090518
ENSRNOT00000083489 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr17_+_36956385 | 0.01 |
ENSRNOT00000024854
|
Cdkal1
|
CDK5 regulatory subunit associated protein 1-like 1 |
| chr6_+_128973956 | 0.01 |
ENSRNOT00000075399
|
Fam181a
|
family with sequence similarity 181, member A |
| chr10_-_44147035 | 0.01 |
ENSRNOT00000041113
|
Olr1424
|
olfactory receptor 1424 |
| chr7_-_143473697 | 0.01 |
ENSRNOT00000055336
ENSRNOT00000083027 |
Krt77
|
keratin 77 |
| chr20_+_5527181 | 0.01 |
ENSRNOT00000091364
|
Phf1
|
PHD finger protein 1 |
| chr8_-_48850671 | 0.01 |
ENSRNOT00000016580
|
Cxcr5
|
C-X-C motif chemokine receptor 5 |
| chr14_-_45165207 | 0.01 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr10_+_104523996 | 0.01 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr1_-_166912524 | 0.00 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr3_-_166994286 | 0.00 |
ENSRNOT00000081593
|
Zfp217
|
zinc finger protein 217 |
| chr1_-_141470380 | 0.00 |
ENSRNOT00000065759
|
Plin1
|
perilipin 1 |
| chrX_+_20216587 | 0.00 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr8_-_40122450 | 0.00 |
ENSRNOT00000039729
|
Tbrg1
|
transforming growth factor beta regulator 1 |
| chr14_+_82769642 | 0.00 |
ENSRNOT00000065393
|
Ctbp1
|
C-terminal binding protein 1 |
| chr5_-_151029233 | 0.00 |
ENSRNOT00000089155
|
Ppp1r8
|
protein phosphatase 1, regulatory subunit 8 |
| chr3_+_33440191 | 0.00 |
ENSRNOT00000034632
ENSRNOT00000092907 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chr10_+_53595854 | 0.00 |
ENSRNOT00000044116
|
Sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr19_+_38189605 | 0.00 |
ENSRNOT00000000275
|
Prmt7
|
protein arginine methyltransferase 7 |
| chr3_-_166993940 | 0.00 |
ENSRNOT00000034669
|
Zfp217
|
zinc finger protein 217 |
| chr17_-_69711689 | 0.00 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.0 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.0 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |