Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
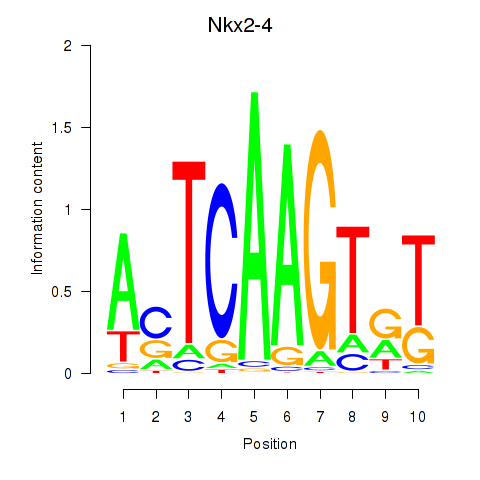
Results for Nkx2-4
Z-value: 0.19
Transcription factors associated with Nkx2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-4
|
ENSRNOG00000012647 | NK2 homeobox 4 |
Activity profile of Nkx2-4 motif
Sorted Z-values of Nkx2-4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_61332391 | 0.14 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr15_-_60289763 | 0.06 |
ENSRNOT00000038579
|
Fam216b
|
family with sequence similarity 216, member B |
| chr16_-_8564379 | 0.06 |
ENSRNOT00000060975
|
LOC680885
|
hypothetical protein LOC680885 |
| chr4_-_160662974 | 0.06 |
ENSRNOT00000089199
|
Tspan9
|
tetraspanin 9 |
| chr7_+_140742418 | 0.06 |
ENSRNOT00000089211
|
Prph
|
peripherin |
| chr14_-_85191557 | 0.05 |
ENSRNOT00000011604
|
Nefh
|
neurofilament heavy |
| chr7_+_136182224 | 0.04 |
ENSRNOT00000008159
|
Tmem117
|
transmembrane protein 117 |
| chr15_-_76789298 | 0.04 |
ENSRNOT00000077853
|
AABR07018857.1
|
|
| chr18_-_63394690 | 0.03 |
ENSRNOT00000090078
ENSRNOT00000029431 |
Cep76
|
centrosomal protein 76 |
| chr17_-_29895386 | 0.03 |
ENSRNOT00000048757
|
Cdyl
|
chromodomain Y-like |
| chr6_+_43001948 | 0.03 |
ENSRNOT00000007374
|
Hpcal1
|
hippocalcin-like 1 |
| chr3_-_9936352 | 0.03 |
ENSRNOT00000011224
ENSRNOT00000042798 |
Fnbp1
|
formin binding protein 1 |
| chr5_+_149077412 | 0.03 |
ENSRNOT00000014666
|
Matn1
|
matrilin 1, cartilage matrix protein |
| chr12_-_19114399 | 0.02 |
ENSRNOT00000073099
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr18_+_29972808 | 0.02 |
ENSRNOT00000074051
|
Pcdha4
|
protocadherin alpha 4 |
| chr3_-_93216495 | 0.02 |
ENSRNOT00000010580
|
Ehf
|
ets homologous factor |
| chr10_+_57606650 | 0.02 |
ENSRNOT00000044363
ENSRNOT00000009082 |
Rpain
|
RPA interacting protein |
| chr13_-_90814119 | 0.02 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr2_+_68821004 | 0.02 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr1_-_242765807 | 0.02 |
ENSRNOT00000020763
|
Pgm5
|
phosphoglucomutase 5 |
| chr4_+_64088900 | 0.02 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr16_-_23903884 | 0.01 |
ENSRNOT00000043305
|
Nat3
|
N-acetyltransferase 3 |
| chr1_-_100671074 | 0.01 |
ENSRNOT00000027132
|
Myh14
|
myosin heavy chain 14 |
| chrX_+_74497262 | 0.01 |
ENSRNOT00000003899
|
Zcchc13
|
zinc finger CCHC-type containing 13 |
| chr7_+_35472102 | 0.01 |
ENSRNOT00000010316
|
Tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr1_-_222224910 | 0.01 |
ENSRNOT00000028720
|
Plcb3
|
phospholipase C beta 3 |
| chr4_-_49422824 | 0.01 |
ENSRNOT00000077889
|
Fam3c
|
family with sequence similarity 3, member C |
| chr8_+_117576788 | 0.01 |
ENSRNOT00000027584
|
Ip6k2
|
inositol hexakisphosphate kinase 2 |
| chr1_+_75233703 | 0.01 |
ENSRNOT00000064178
|
LOC100911727
|
DNA ligase 1-like |
| chr1_+_185863043 | 0.01 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr14_-_14390699 | 0.01 |
ENSRNOT00000046639
|
Anxa3
|
annexin A3 |
| chr5_+_2632712 | 0.01 |
ENSRNOT00000009431
|
Rpl7
|
ribosomal protein L7 |
| chr2_+_195996521 | 0.01 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr1_-_44615760 | 0.01 |
ENSRNOT00000022148
|
Nox3
|
NADPH oxidase 3 |
| chr2_-_235177275 | 0.01 |
ENSRNOT00000093153
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr2_-_193335002 | 0.01 |
ENSRNOT00000012691
|
Lce1m
|
late cornified envelope 1M |
| chr12_-_2545740 | 0.01 |
ENSRNOT00000075527
|
Tgfbr3l
|
transforming growth factor beta receptor 3 like |
| chr5_-_2632030 | 0.01 |
ENSRNOT00000009096
|
Rdh10
|
retinol dehydrogenase 10 |
| chr8_+_119382011 | 0.01 |
ENSRNOT00000056060
|
Lrrfip2
|
LRR binding FLII interacting protein 2 |
| chr5_-_62260089 | 0.01 |
ENSRNOT00000064858
ENSRNOT00000011714 |
Tbc1d2
|
TBC1 domain family, member 2 |
| chr14_+_72889038 | 0.01 |
ENSRNOT00000084261
|
Rab5a
|
RAB5A, member RAS oncogene family |
| chr2_+_68820615 | 0.01 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr4_+_82783686 | 0.01 |
ENSRNOT00000041988
|
Tax1bp1
|
Tax1 binding protein 1 |
| chr1_+_170147300 | 0.01 |
ENSRNOT00000048075
|
Olr200
|
olfactory receptor 200 |
| chr6_-_128434183 | 0.01 |
ENSRNOT00000014405
|
Dicer1
|
dicer 1 ribonuclease III |
| chr6_+_10306405 | 0.01 |
ENSRNOT00000090420
ENSRNOT00000080507 |
Epas1
|
endothelial PAS domain protein 1 |
| chr9_+_95221474 | 0.00 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr2_+_95320283 | 0.00 |
ENSRNOT00000015537
|
Hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr15_-_14510828 | 0.00 |
ENSRNOT00000039347
|
Sntn
|
sentan, cilia apical structure protein |
| chr8_-_55162421 | 0.00 |
ENSRNOT00000083102
|
Dixdc1
|
DIX domain containing 1 |
| chr18_+_63394900 | 0.00 |
ENSRNOT00000024126
|
Psmg2
|
proteasome assembly chaperone 2 |
| chr4_-_129430251 | 0.00 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr1_+_141821916 | 0.00 |
ENSRNOT00000071574
|
Echs1
|
enoyl-CoA hydratase, short chain 1 |
| chr2_-_192780631 | 0.00 |
ENSRNOT00000012371
|
Smcp
|
sperm mitochondria-associated cysteine-rich protein |
| chr16_-_82099991 | 0.00 |
ENSRNOT00000089447
|
Atp11a
|
ATPase phospholipid transporting 11A |
| chr1_-_212579057 | 0.00 |
ENSRNOT00000025446
|
LOC100911186
|
enoyl-CoA hydratase, mitochondrial-like |
| chr2_+_4989295 | 0.00 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chrX_+_33528081 | 0.00 |
ENSRNOT00000006584
|
Syap1
|
synapse associated protein 1 |
| chr1_-_67134827 | 0.00 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr12_-_13133219 | 0.00 |
ENSRNOT00000092292
|
Daglb
|
diacylglycerol lipase, beta |
| chr16_-_55132191 | 0.00 |
ENSRNOT00000017025
|
Micu3
|
mitochondrial calcium uptake family, member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |