Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
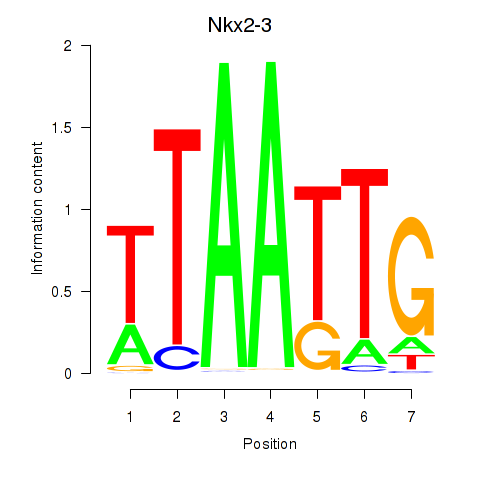
Results for Nkx2-3
Z-value: 0.54
Transcription factors associated with Nkx2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-3
|
ENSRNOG00000016656 | NK2 homeobox 3 |
Activity profile of Nkx2-3 motif
Sorted Z-values of Nkx2-3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_56766475 | 0.25 |
ENSRNOT00000078819
|
Sp5
|
Sp5 transcription factor |
| chr7_+_42304534 | 0.17 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr2_+_41467064 | 0.16 |
ENSRNOT00000073231
|
AABR07008066.2
|
|
| chr13_+_87986240 | 0.15 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr1_+_256382791 | 0.12 |
ENSRNOT00000022549
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr2_-_138833933 | 0.11 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chr9_+_25410669 | 0.11 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr7_+_41475163 | 0.10 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr9_-_30844199 | 0.10 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr9_-_121725716 | 0.10 |
ENSRNOT00000087405
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr7_-_122644054 | 0.09 |
ENSRNOT00000025941
|
Dnajb7
|
DnaJ heat shock protein family (Hsp40) member B7 |
| chr6_+_58468155 | 0.09 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr3_-_141411170 | 0.09 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr2_-_45518502 | 0.09 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr10_-_87286387 | 0.09 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr1_-_166912524 | 0.09 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr1_-_275882444 | 0.08 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr11_+_82848853 | 0.08 |
ENSRNOT00000073743
|
Thpo
|
thrombopoietin |
| chrX_+_158835811 | 0.08 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr17_-_43675934 | 0.08 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr6_-_106971250 | 0.08 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr4_+_96562725 | 0.08 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr3_-_45169118 | 0.07 |
ENSRNOT00000086371
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr2_-_96509424 | 0.07 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chrX_+_13441558 | 0.07 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr5_+_25349928 | 0.07 |
ENSRNOT00000020826
|
Gem
|
GTP binding protein overexpressed in skeletal muscle |
| chr2_-_5577369 | 0.07 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr6_+_106496992 | 0.07 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr15_+_57505449 | 0.07 |
ENSRNOT00000064945
|
Siah3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr14_-_16903242 | 0.07 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr13_-_32427177 | 0.06 |
ENSRNOT00000044628
|
Cdh19
|
cadherin 19 |
| chr9_+_52023295 | 0.06 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr17_-_51912496 | 0.06 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr1_+_53220397 | 0.06 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr13_+_47935163 | 0.06 |
ENSRNOT00000006768
|
Eif2d
|
eukaryotic translation initiation factor 2D |
| chr7_+_125649688 | 0.06 |
ENSRNOT00000017906
|
Phf21b
|
PHD finger protein 21B |
| chr15_+_37790141 | 0.06 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr5_+_147476221 | 0.06 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr7_+_28414350 | 0.06 |
ENSRNOT00000085680
|
Igf1
|
insulin-like growth factor 1 |
| chr12_+_2140203 | 0.06 |
ENSRNOT00000084906
|
Camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr5_-_130085838 | 0.06 |
ENSRNOT00000035252
|
Elavl4
|
ELAV like RNA binding protein 4 |
| chr5_-_151895016 | 0.06 |
ENSRNOT00000081841
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr5_+_119727839 | 0.06 |
ENSRNOT00000014354
|
Cachd1
|
cache domain containing 1 |
| chr2_+_154580558 | 0.05 |
ENSRNOT00000076976
|
Gmps
|
guanine monophosphate synthase |
| chr11_-_768644 | 0.05 |
ENSRNOT00000090197
|
Epha3
|
Eph receptor A3 |
| chr14_+_104250617 | 0.05 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr9_+_47281961 | 0.05 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chrX_-_62690806 | 0.05 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr1_+_261802223 | 0.05 |
ENSRNOT00000081765
|
R3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr19_-_49510901 | 0.05 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr3_+_177310753 | 0.05 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr11_-_45124423 | 0.05 |
ENSRNOT00000046383
|
Filip1l
|
filamin A interacting protein 1-like |
| chr9_+_14529218 | 0.05 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr4_+_174810959 | 0.05 |
ENSRNOT00000035507
|
Aebp2
|
AE binding protein 2 |
| chr13_-_90977734 | 0.04 |
ENSRNOT00000011869
|
Slamf8
|
SLAM family member 8 |
| chr3_+_168124673 | 0.04 |
ENSRNOT00000070809
|
Pfdn4
|
prefoldin subunit 4 |
| chr8_-_82533689 | 0.04 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr10_-_4257868 | 0.04 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr1_-_101198801 | 0.04 |
ENSRNOT00000037998
|
Ccdc155
|
coiled-coil domain containing 155 |
| chr11_-_90406797 | 0.04 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr3_+_59981959 | 0.04 |
ENSRNOT00000030460
|
Sp9
|
Sp9 transcription factor |
| chr5_+_173288447 | 0.04 |
ENSRNOT00000091205
ENSRNOT00000067252 |
Mxra8
|
matrix remodeling associated 8 |
| chr1_-_101095594 | 0.04 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr3_+_44025300 | 0.04 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr6_+_2216623 | 0.04 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr18_+_44716226 | 0.04 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chrX_+_54062935 | 0.04 |
ENSRNOT00000004854
|
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr7_-_15027933 | 0.04 |
ENSRNOT00000006697
|
Zfp871
|
zinc finger protein 871 |
| chr5_-_155709215 | 0.04 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chr8_-_94352246 | 0.04 |
ENSRNOT00000013244
|
Me1
|
malic enzyme 1 |
| chr6_+_76745981 | 0.04 |
ENSRNOT00000076784
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr4_-_32392007 | 0.04 |
ENSRNOT00000014946
|
Dlx5
|
distal-less homeobox 5 |
| chr6_-_114476723 | 0.04 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr10_-_67285617 | 0.04 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr6_+_56846789 | 0.04 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr17_-_23792353 | 0.04 |
ENSRNOT00000019089
|
Tbc1d7
|
TBC1 domain family, member 7 |
| chr19_+_22699808 | 0.04 |
ENSRNOT00000023169
|
RGD1308706
|
similar to RIKEN cDNA 4921524J17 |
| chr13_-_88307988 | 0.03 |
ENSRNOT00000003812
|
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chrX_-_64908682 | 0.03 |
ENSRNOT00000084107
|
Zc4h2
|
zinc finger C4H2-type containing |
| chr2_-_185852759 | 0.03 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr11_-_29710849 | 0.03 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chrM_+_7758 | 0.03 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chrX_-_25590048 | 0.03 |
ENSRNOT00000004873
|
Mid1
|
midline 1 |
| chr15_-_88670349 | 0.03 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr4_+_65637407 | 0.03 |
ENSRNOT00000047954
|
Trim24
|
tripartite motif-containing 24 |
| chr17_-_9762813 | 0.03 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr3_-_11452529 | 0.03 |
ENSRNOT00000020206
|
Slc25a25
|
solute carrier family 25 member 25 |
| chr9_-_16519144 | 0.03 |
ENSRNOT00000072581
|
LOC681367
|
hypothetical protein LOC681367 |
| chr1_-_73399579 | 0.03 |
ENSRNOT00000077186
|
Lilrb4
|
leukocyte immunoglobulin like receptor B4 |
| chr14_+_99529284 | 0.03 |
ENSRNOT00000006897
ENSRNOT00000067134 |
Vstm2a
|
V-set and transmembrane domain containing 2A |
| chr3_-_5510908 | 0.03 |
ENSRNOT00000042289
|
Rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr11_-_782954 | 0.03 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr10_+_95770154 | 0.03 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chrX_+_131381134 | 0.03 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr2_-_199354793 | 0.03 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr15_+_70790125 | 0.03 |
ENSRNOT00000012440
|
Tdrd3
|
tudor domain containing 3 |
| chr1_-_173764246 | 0.03 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr4_+_144192989 | 0.03 |
ENSRNOT00000007523
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chrX_+_114930457 | 0.03 |
ENSRNOT00000089141
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr9_-_63641400 | 0.03 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr1_+_217459215 | 0.03 |
ENSRNOT00000092386
ENSRNOT00000092357 |
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_+_52127632 | 0.03 |
ENSRNOT00000079797
|
Cadm1
|
cell adhesion molecule 1 |
| chr7_-_134560713 | 0.03 |
ENSRNOT00000006621
|
Yaf2
|
YY1 associated factor 2 |
| chrX_-_40086870 | 0.03 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr14_+_108826831 | 0.03 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr8_-_53816447 | 0.03 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr5_+_164845925 | 0.03 |
ENSRNOT00000011384
|
Mthfr
|
methylenetetrahydrofolate reductase |
| chr12_-_9864791 | 0.03 |
ENSRNOT00000073026
|
Gtf3a
|
general transcription factor III A |
| chr2_+_186980793 | 0.03 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chrX_-_10031167 | 0.03 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr7_+_133856101 | 0.03 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr8_+_12994155 | 0.03 |
ENSRNOT00000011855
|
Cwc15
|
CWC15 spliceosome-associated protein |
| chr9_+_25304876 | 0.03 |
ENSRNOT00000065053
|
Tfap2d
|
transcription factor AP-2 delta |
| chr17_+_11683862 | 0.03 |
ENSRNOT00000024766
|
Msx2
|
msh homeobox 2 |
| chr14_-_79988110 | 0.03 |
ENSRNOT00000090597
|
Afap1
|
actin filament associated protein 1 |
| chr15_-_109316953 | 0.03 |
ENSRNOT00000019158
|
Ggact
|
gamma-glutamylamine cyclotransferase |
| chr7_+_122160171 | 0.03 |
ENSRNOT00000074499
|
LOC100362980
|
CG3918-like |
| chr5_-_99033107 | 0.02 |
ENSRNOT00000041306
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chrX_-_118998300 | 0.02 |
ENSRNOT00000025575
|
AABR07041089.1
|
|
| chr12_+_37603986 | 0.02 |
ENSRNOT00000088006
|
Sbno1
|
strawberry notch homolog 1 |
| chr1_+_1784078 | 0.02 |
ENSRNOT00000020098
|
Lats1
|
large tumor suppressor kinase 1 |
| chr1_-_80615704 | 0.02 |
ENSRNOT00000041891
|
Apoe
|
apolipoprotein E |
| chrX_-_42329232 | 0.02 |
ENSRNOT00000045705
|
LOC100361934
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 |
| chr6_-_51018050 | 0.02 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr8_+_52127399 | 0.02 |
ENSRNOT00000052392
|
Cadm1
|
cell adhesion molecule 1 |
| chr10_+_82032656 | 0.02 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr10_+_94260197 | 0.02 |
ENSRNOT00000063973
|
Taco1
|
translational activator of cytochrome c oxidase I |
| chr4_-_178168690 | 0.02 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr5_+_24410863 | 0.02 |
ENSRNOT00000010591
|
Tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr6_-_67084234 | 0.02 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr18_+_30424814 | 0.02 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chr2_+_144861455 | 0.02 |
ENSRNOT00000093284
ENSRNOT00000019748 ENSRNOT00000072110 |
Dclk1
|
doublecortin-like kinase 1 |
| chr10_+_84147836 | 0.02 |
ENSRNOT00000010527
|
Hoxb6
|
homeo box B6 |
| chr8_+_100260049 | 0.02 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr1_-_15374850 | 0.02 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr2_-_140464607 | 0.02 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
| chr4_-_28437676 | 0.02 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr16_-_6245644 | 0.02 |
ENSRNOT00000040759
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr12_+_37709860 | 0.02 |
ENSRNOT00000086244
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr9_-_66019065 | 0.02 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr17_-_81187739 | 0.02 |
ENSRNOT00000063911
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr5_+_98387291 | 0.02 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr9_-_71798265 | 0.02 |
ENSRNOT00000043766
|
Crygb
|
crystallin, gamma B |
| chr14_+_108415068 | 0.02 |
ENSRNOT00000083763
|
Pus10
|
pseudouridylate synthase 10 |
| chr8_+_85489553 | 0.02 |
ENSRNOT00000082158
|
Rpl12-ps1
|
ribosomal protein L12, pseudogene 1 |
| chr1_+_22332090 | 0.02 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr11_-_60546997 | 0.02 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr6_+_73553210 | 0.02 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr18_+_73256085 | 0.02 |
ENSRNOT00000080660
|
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chrX_+_33443186 | 0.02 |
ENSRNOT00000005622
|
S100g
|
S100 calcium binding protein G |
| chr4_+_8066907 | 0.02 |
ENSRNOT00000080811
|
Srpk2
|
SRSF protein kinase 2 |
| chr7_-_95310005 | 0.02 |
ENSRNOT00000005815
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr14_+_108143869 | 0.02 |
ENSRNOT00000090003
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr14_-_21748356 | 0.02 |
ENSRNOT00000002670
|
Cabs1
|
calcium binding protein, spermatid associated 1 |
| chr3_+_113918629 | 0.02 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr2_+_154604832 | 0.02 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chrX_-_26376467 | 0.02 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr20_+_25990656 | 0.02 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr15_-_37831031 | 0.02 |
ENSRNOT00000091562
|
Eef1akmt1
|
eukaryotic translation elongation factor 1 alpha lysine methyltransferase 1 |
| chr14_-_8510138 | 0.02 |
ENSRNOT00000080758
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr14_+_37130743 | 0.02 |
ENSRNOT00000031756
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr4_-_15859132 | 0.02 |
ENSRNOT00000082161
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr1_+_224998172 | 0.02 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr15_+_59678165 | 0.02 |
ENSRNOT00000074868
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr7_+_72924799 | 0.02 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr1_-_198794947 | 0.02 |
ENSRNOT00000024639
|
Znf768
|
zinc finger protein 768 |
| chr6_+_69971227 | 0.02 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr4_+_8066737 | 0.02 |
ENSRNOT00000063994
|
Srpk2
|
SRSF protein kinase 2 |
| chr13_-_90405591 | 0.02 |
ENSRNOT00000006849
|
Vangl2
|
VANGL planar cell polarity protein 2 |
| chr8_+_44990014 | 0.02 |
ENSRNOT00000044608
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr11_+_80736576 | 0.02 |
ENSRNOT00000047678
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr6_+_28382962 | 0.02 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr7_-_107391184 | 0.02 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr2_+_204427608 | 0.02 |
ENSRNOT00000083374
|
Nhlh2
|
nescient helix loop helix 2 |
| chr2_-_187258140 | 0.02 |
ENSRNOT00000017735
|
Prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr12_-_44520341 | 0.02 |
ENSRNOT00000066810
|
Nos1
|
nitric oxide synthase 1 |
| chr4_+_68656928 | 0.02 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr4_-_113497396 | 0.02 |
ENSRNOT00000079173
ENSRNOT00000067314 |
Pole4
|
DNA polymerase epsilon 4, accessory subunit |
| chr7_-_60416925 | 0.02 |
ENSRNOT00000008193
|
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr14_-_19072677 | 0.02 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr7_-_142063212 | 0.02 |
ENSRNOT00000089912
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr13_+_98231326 | 0.01 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr2_-_123972356 | 0.01 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr20_+_248410 | 0.01 |
ENSRNOT00000049573
|
Tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr12_+_38377034 | 0.01 |
ENSRNOT00000088002
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr7_-_55604403 | 0.01 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chrX_+_113510621 | 0.01 |
ENSRNOT00000025912
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr13_-_102857551 | 0.01 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr8_+_106449321 | 0.01 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr9_-_18371856 | 0.01 |
ENSRNOT00000027293
|
Supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr13_-_111972603 | 0.01 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr18_+_29999290 | 0.01 |
ENSRNOT00000027372
|
Pcdha4
|
protocadherin alpha 4 |
| chr8_-_21492801 | 0.01 |
ENSRNOT00000077465
|
Zfp426
|
zinc finger protein 426 |
| chr20_+_26999795 | 0.01 |
ENSRNOT00000057872
|
Mypn
|
myopalladin |
| chr10_+_110406450 | 0.01 |
ENSRNOT00000054924
|
Hexdc
|
hexosaminidase D |
| chr2_+_186980992 | 0.01 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr2_+_145174876 | 0.01 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr18_-_24182012 | 0.01 |
ENSRNOT00000023594
|
Syt4
|
synaptotagmin 4 |
| chr1_+_228337767 | 0.01 |
ENSRNOT00000066247
|
Patl1
|
PAT1 homolog 1, processing body mRNA decay factor |
| chr4_-_184096806 | 0.01 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070662 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:1901367 | response to L-cysteine(GO:1901367) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.0 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 0.0 | 0.0 | GO:0090172 | microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.0 | GO:0071338 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.0 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine to uridine editing(GO:0016554) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.0 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.0 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0003921 | GMP synthase activity(GO:0003921) GMP synthase (glutamine-hydrolyzing) activity(GO:0003922) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |