Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
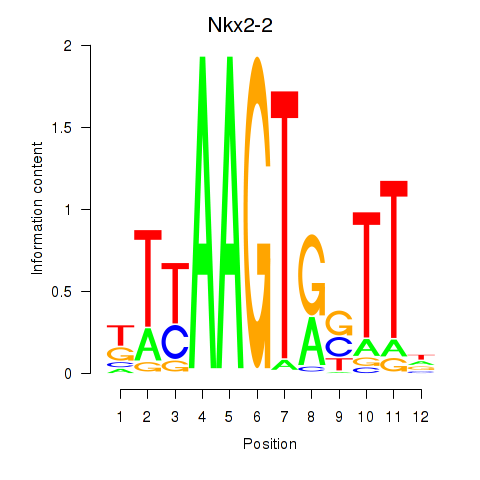
Results for Nkx2-2
Z-value: 0.42
Transcription factors associated with Nkx2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-2
|
ENSRNOG00000012728 | NK2 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-2 | rn6_v1_chr3_-_141411170_141411170 | -0.62 | 2.7e-01 | Click! |
Activity profile of Nkx2-2 motif
Sorted Z-values of Nkx2-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_28654733 | 0.89 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr14_-_37770059 | 0.15 |
ENSRNOT00000029721
|
Slc10a4
|
solute carrier family 10, member 4 |
| chr2_+_71753816 | 0.14 |
ENSRNOT00000025809
|
AABR07008724.1
|
|
| chr20_-_22459025 | 0.12 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr14_+_88549947 | 0.11 |
ENSRNOT00000086177
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr4_-_120414118 | 0.11 |
ENSRNOT00000072795
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr10_-_13780283 | 0.10 |
ENSRNOT00000084200
|
AC103090.1
|
|
| chr1_-_227047290 | 0.10 |
ENSRNOT00000037226
|
Ms4a15
|
membrane spanning 4-domains A15 |
| chr4_-_129430251 | 0.10 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr4_+_6903198 | 0.10 |
ENSRNOT00000012285
|
Crygn
|
crystallin, gamma N |
| chr8_+_82288705 | 0.10 |
ENSRNOT00000012409
|
Bcl2l10
|
BCL2 like 10 |
| chr6_+_133552821 | 0.09 |
ENSRNOT00000006339
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr20_-_4366957 | 0.08 |
ENSRNOT00000088244
|
Rnf5
|
ring finger protein 5 |
| chr20_-_48310175 | 0.08 |
ENSRNOT00000000351
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr6_+_3375476 | 0.08 |
ENSRNOT00000045349
|
LOC102551744
|
eukaryotic translation initiation factor 1-like |
| chr17_+_27452638 | 0.08 |
ENSRNOT00000038349
|
Cage1
|
cancer antigen 1 |
| chr10_+_14258380 | 0.08 |
ENSRNOT00000021650
|
Nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr5_-_155709215 | 0.08 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chr12_+_37668369 | 0.07 |
ENSRNOT00000001419
|
Cdk2ap1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr9_+_94928489 | 0.07 |
ENSRNOT00000087366
ENSRNOT00000085770 ENSRNOT00000024733 |
Sag
|
S-antigen visual arrestin |
| chr9_-_121972055 | 0.07 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr2_+_207350651 | 0.07 |
ENSRNOT00000019129
|
St7l
|
suppression of tumorigenicity 7-like |
| chr5_-_61077627 | 0.07 |
ENSRNOT00000015338
|
Shb
|
SH2 domain containing adaptor protein B |
| chr14_-_44375804 | 0.07 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr5_+_10125070 | 0.07 |
ENSRNOT00000085321
|
AABR07046866.1
|
|
| chr14_-_43992587 | 0.07 |
ENSRNOT00000003425
|
Rhoh
|
ras homolog family member H |
| chr4_-_28437676 | 0.07 |
ENSRNOT00000012995
|
Hepacam2
|
HEPACAM family member 2 |
| chr18_-_86279680 | 0.07 |
ENSRNOT00000006169
|
LOC689166
|
hypothetical protein LOC689166 |
| chr17_+_44763598 | 0.07 |
ENSRNOT00000079880
|
Hist1h3b
|
histone cluster 1, H3b |
| chr1_+_227670159 | 0.07 |
ENSRNOT00000072077
|
Ms4a6c
|
membrane-spanning 4-domains, subfamily A, member 6C |
| chr14_-_18531179 | 0.06 |
ENSRNOT00000003703
|
Areg
|
amphiregulin |
| chr5_+_144031402 | 0.06 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr6_+_8669722 | 0.06 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr17_+_85500113 | 0.06 |
ENSRNOT00000091937
|
AABR07028768.1
|
|
| chr18_-_74151950 | 0.06 |
ENSRNOT00000023101
|
Haus1
|
HAUS augmin-like complex, subunit 1 |
| chr1_+_169145445 | 0.06 |
ENSRNOT00000034019
|
LOC499219
|
hypothetical protein LOC499219 |
| chr2_-_98610368 | 0.06 |
ENSRNOT00000011641
|
Zfhx4
|
zinc finger homeobox 4 |
| chr10_+_40247436 | 0.06 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr13_-_52136127 | 0.06 |
ENSRNOT00000009398
|
Timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr4_-_120041238 | 0.06 |
ENSRNOT00000073799
|
LOC100911337
|
40S ribosomal protein S25-like |
| chr1_+_98398660 | 0.06 |
ENSRNOT00000047473
|
Cd33
|
CD33 molecule |
| chr8_+_79054237 | 0.06 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr3_+_149466770 | 0.06 |
ENSRNOT00000065104
|
Bpifa6
|
BPI fold containing family A, member 6 |
| chr19_+_56220755 | 0.06 |
ENSRNOT00000023452
|
Tubb3
|
tubulin, beta 3 class III |
| chr20_+_13827132 | 0.06 |
ENSRNOT00000001664
|
Ddt
|
D-dopachrome tautomerase |
| chr14_+_88543630 | 0.06 |
ENSRNOT00000060515
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr20_-_4542947 | 0.06 |
ENSRNOT00000086140
|
Cfb
|
complement factor B |
| chr2_+_207351051 | 0.06 |
ENSRNOT00000084302
|
St7l
|
suppression of tumorigenicity 7-like |
| chr7_-_29949603 | 0.06 |
ENSRNOT00000009419
|
LOC103692829
|
60S ribosomal protein L9 pseudogene |
| chr1_+_175445088 | 0.06 |
ENSRNOT00000036718
|
Adm
|
adrenomedullin |
| chr1_+_101410019 | 0.05 |
ENSRNOT00000042039
|
Lhb
|
luteinizing hormone beta polypeptide |
| chr1_-_104202591 | 0.05 |
ENSRNOT00000035512
|
E2f8
|
E2F transcription factor 8 |
| chr7_-_68512397 | 0.05 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr10_+_14062331 | 0.05 |
ENSRNOT00000029652
|
Noxo1
|
NADPH oxidase organizer 1 |
| chr1_-_211956858 | 0.05 |
ENSRNOT00000054886
|
Cfap46
|
cilia and flagella associated protein 46 |
| chr13_-_89433815 | 0.05 |
ENSRNOT00000091541
|
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr10_-_39405311 | 0.05 |
ENSRNOT00000074616
|
Pdlim4
|
PDZ and LIM domain 4 |
| chr1_-_80716143 | 0.05 |
ENSRNOT00000092048
ENSRNOT00000025829 |
Cblc
|
Cbl proto-oncogene C |
| chr1_-_44474674 | 0.05 |
ENSRNOT00000078768
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr15_+_4850122 | 0.05 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chrX_+_78259409 | 0.05 |
ENSRNOT00000049779
|
RGD1560455
|
similar to RIKEN cDNA A630033H20 gene |
| chr20_-_4391402 | 0.05 |
ENSRNOT00000090518
ENSRNOT00000083489 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr6_-_22138286 | 0.05 |
ENSRNOT00000007607
|
Yipf4
|
Yip1 domain family, member 4 |
| chr1_-_233140237 | 0.05 |
ENSRNOT00000083372
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr1_-_196883051 | 0.05 |
ENSRNOT00000055034
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr2_-_154763094 | 0.05 |
ENSRNOT00000031015
|
Vom2r46
|
vomeronasal 2 receptor, 46 |
| chr19_-_9843673 | 0.05 |
ENSRNOT00000015795
|
Gins3
|
GINS complex subunit 3 |
| chr1_+_7252349 | 0.05 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr11_+_32450587 | 0.05 |
ENSRNOT00000061235
|
Smim11
|
small integral membrane protein 11 |
| chr2_-_91497091 | 0.05 |
ENSRNOT00000015185
|
Pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr2_-_147392062 | 0.05 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr20_+_47911018 | 0.05 |
ENSRNOT00000038842
|
LOC688390
|
hypothetical protein LOC688390 |
| chr2_-_26870421 | 0.05 |
ENSRNOT00000088791
|
AABR07007744.2
|
|
| chr18_+_81694808 | 0.05 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chrX_-_139229702 | 0.05 |
ENSRNOT00000042507
|
AABR07041771.1
|
|
| chr10_+_85257876 | 0.04 |
ENSRNOT00000014752
|
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chr1_-_98570949 | 0.04 |
ENSRNOT00000033648
|
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chrX_+_110404022 | 0.04 |
ENSRNOT00000041044
|
Mum1l1
|
MUM1 like 1 |
| chr17_-_13393243 | 0.04 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr17_-_43537293 | 0.04 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chrX_-_138435391 | 0.04 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
| chr16_-_38063821 | 0.04 |
ENSRNOT00000077452
ENSRNOT00000089533 |
Glra3
|
glycine receptor, alpha 3 |
| chr16_-_37908173 | 0.04 |
ENSRNOT00000075026
|
Glra3
|
glycine receptor, alpha 3 |
| chrX_+_14019961 | 0.04 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr18_+_30023828 | 0.04 |
ENSRNOT00000079008
|
Pcdha4
|
protocadherin alpha 4 |
| chr5_-_164747083 | 0.04 |
ENSRNOT00000010433
|
Plod1
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chrX_+_78372257 | 0.04 |
ENSRNOT00000046164
|
Gpr174
|
G protein-coupled receptor 174 |
| chr5_-_151895016 | 0.04 |
ENSRNOT00000081841
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr2_+_92559929 | 0.04 |
ENSRNOT00000033404
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr7_+_107413691 | 0.04 |
ENSRNOT00000007587
|
Phf20l1
|
PHD finger protein 20-like protein 1 |
| chr7_-_54778848 | 0.04 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr3_+_15379109 | 0.04 |
ENSRNOT00000036495
|
Morn5
|
MORN repeat containing 5 |
| chr14_+_7630482 | 0.04 |
ENSRNOT00000091581
|
LOC498330
|
similar to hypothetical protein MGC26744 |
| chr3_+_112950890 | 0.04 |
ENSRNOT00000015472
|
Ccndbp1
|
cyclin D1 binding protein 1 |
| chr13_-_90814119 | 0.04 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr1_-_163148948 | 0.04 |
ENSRNOT00000046850
|
B3gnt6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 |
| chr1_+_93242050 | 0.04 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr18_+_35574002 | 0.04 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr9_+_95221474 | 0.04 |
ENSRNOT00000066839
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr18_+_57654290 | 0.04 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr20_-_5112258 | 0.04 |
ENSRNOT00000092222
|
Csnk2b
|
casein kinase 2 beta |
| chr7_+_129973480 | 0.04 |
ENSRNOT00000050620
ENSRNOT00000042686 |
Mov10l1
|
Mov10 RISC complex RNA helicase like 1 |
| chr4_-_182600258 | 0.04 |
ENSRNOT00000067970
|
Ergic2
|
ERGIC and golgi 2 |
| chr8_-_132919110 | 0.04 |
ENSRNOT00000008747
|
Xcr1
|
X-C motif chemokine receptor 1 |
| chr8_-_39266959 | 0.04 |
ENSRNOT00000046590
|
Ei24
|
EI24, autophagy associated transmembrane protein |
| chr7_+_140781799 | 0.04 |
ENSRNOT00000087932
|
Dnajc22
|
DnaJ heat shock protein family (Hsp40) member C22 |
| chr12_-_22710674 | 0.04 |
ENSRNOT00000050456
|
Mogat3
|
monoacylglycerol O-acyltransferase 3 |
| chr13_+_42220251 | 0.04 |
ENSRNOT00000078185
|
AABR07020815.1
|
|
| chr6_+_129609375 | 0.04 |
ENSRNOT00000083732
|
Papola
|
poly (A) polymerase alpha |
| chr2_-_93641497 | 0.04 |
ENSRNOT00000013720
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr3_-_133232322 | 0.04 |
ENSRNOT00000034487
|
Esf1
|
ESF1 nucleolar pre-rRNA processing protein homolog |
| chr10_-_56429748 | 0.04 |
ENSRNOT00000020675
ENSRNOT00000092704 |
Spem1
|
spermatid maturation 1 |
| chr3_-_120011364 | 0.04 |
ENSRNOT00000018922
|
Fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr8_-_115149419 | 0.04 |
ENSRNOT00000016056
ENSRNOT00000077514 |
Abhd14a
|
abhydrolase domain containing 14A |
| chr19_+_26022849 | 0.03 |
ENSRNOT00000014887
|
Dnase2
|
deoxyribonuclease 2, lysosomal |
| chr14_-_11198194 | 0.03 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr11_+_74984613 | 0.03 |
ENSRNOT00000035049
|
Atp13a5
|
ATPase 13A5 |
| chr4_-_140220487 | 0.03 |
ENSRNOT00000009008
|
Sumf1
|
sulfatase modifying factor 1 |
| chr5_+_15043955 | 0.03 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chr9_+_37727942 | 0.03 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr13_+_90116843 | 0.03 |
ENSRNOT00000006306
|
Cd48
|
Cd48 molecule |
| chr1_+_102924059 | 0.03 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr9_+_91001828 | 0.03 |
ENSRNOT00000080956
|
AABR07068214.1
|
|
| chr10_+_58613674 | 0.03 |
ENSRNOT00000010975
|
Fam64a
|
family with sequence similarity 64, member A |
| chr10_-_66848388 | 0.03 |
ENSRNOT00000018891
|
Omg
|
oligodendrocyte-myelin glycoprotein |
| chr11_-_78327282 | 0.03 |
ENSRNOT00000002636
|
Tp63
|
tumor protein p63 |
| chr2_+_205568935 | 0.03 |
ENSRNOT00000025248
|
Ampd1
|
adenosine monophosphate deaminase 1 |
| chr4_-_98593664 | 0.03 |
ENSRNOT00000007927
|
Rpia
|
ribose 5-phosphate isomerase A |
| chr9_-_71852113 | 0.03 |
ENSRNOT00000083263
ENSRNOT00000072983 |
AABR07067896.1
|
|
| chr1_+_125229469 | 0.03 |
ENSRNOT00000021884
|
Mcee
|
methylmalonyl CoA epimerase |
| chr14_+_100135271 | 0.03 |
ENSRNOT00000032965
|
Fbxo48
|
F-box protein 48 |
| chr8_+_99636749 | 0.03 |
ENSRNOT00000086524
|
Plscr1
|
phospholipid scramblase 1 |
| chr1_+_101623308 | 0.03 |
ENSRNOT00000029167
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr13_+_51218468 | 0.03 |
ENSRNOT00000033636
|
LOC289035
|
similar to UDP-N-acetylglucosamine:a-1,3-D-mannoside beta-1,4-N-acetylgluco |
| chrX_-_113228265 | 0.03 |
ENSRNOT00000060001
|
RGD1566136
|
similar to 40S ribosomal protein S9 |
| chr2_-_250923744 | 0.03 |
ENSRNOT00000084996
|
Clca1
|
chloride channel accessory 1 |
| chr1_-_190965115 | 0.03 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr1_+_169562247 | 0.03 |
ENSRNOT00000023124
|
Olr153
|
olfactory receptor 153 |
| chr1_+_101249522 | 0.03 |
ENSRNOT00000033882
|
Slc6a16
|
solute carrier family 6, member 16 |
| chr9_+_92681078 | 0.03 |
ENSRNOT00000034152
|
Sp100
|
SP100 nuclear antigen |
| chr7_-_104556419 | 0.03 |
ENSRNOT00000090784
|
Fam49b
|
family with sequence similarity 49, member B |
| chr13_-_89874008 | 0.03 |
ENSRNOT00000051368
|
Ptma
|
prothymosin alpha |
| chr10_-_87437455 | 0.03 |
ENSRNOT00000051080
|
Krt39
|
keratin 39 |
| chr12_-_2826378 | 0.03 |
ENSRNOT00000061749
|
Clec4m
|
C-type lectin domain family 4 member M |
| chr5_-_6083083 | 0.03 |
ENSRNOT00000072411
|
Sulf1
|
sulfatase 1 |
| chr8_-_108777717 | 0.03 |
ENSRNOT00000034188
|
Il20rb
|
interleukin 20 receptor subunit beta |
| chr13_+_52887649 | 0.03 |
ENSRNOT00000048033
|
Ascl5
|
achaete-scute family bHLH transcription factor 5 |
| chr12_-_19439977 | 0.03 |
ENSRNOT00000060035
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr8_+_113603533 | 0.03 |
ENSRNOT00000017280
|
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr13_+_76962504 | 0.03 |
ENSRNOT00000076581
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr16_-_32439421 | 0.03 |
ENSRNOT00000043100
|
Nek1
|
NIMA-related kinase 1 |
| chr7_+_133856101 | 0.03 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr4_+_97657671 | 0.03 |
ENSRNOT00000075400
|
Gng12
|
G protein subunit gamma 12 |
| chr3_+_133232432 | 0.03 |
ENSRNOT00000006412
|
Ndufaf5
|
NADH:ubiquinone oxidoreductase complex assembly factor 5 |
| chr14_-_44225713 | 0.03 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr16_-_20873344 | 0.03 |
ENSRNOT00000027381
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr8_+_55603968 | 0.03 |
ENSRNOT00000066848
|
Pou2af1
|
POU class 2 associating factor 1 |
| chr3_+_160467552 | 0.03 |
ENSRNOT00000066657
|
Stk4
|
serine/threonine kinase 4 |
| chr14_+_33247274 | 0.03 |
ENSRNOT00000075226
|
Spink2
|
serine peptidase inhibitor, Kazal type 2 |
| chr1_+_173252058 | 0.03 |
ENSRNOT00000073421
|
LOC499229
|
similar to very large inducible GTPase 1 isoform A |
| chr1_+_16478127 | 0.03 |
ENSRNOT00000019076
|
Ahi1
|
Abelson helper integration site 1 |
| chr7_+_15621989 | 0.02 |
ENSRNOT00000051928
|
Olr1095
|
olfactory receptor 1095 |
| chr3_+_11476883 | 0.02 |
ENSRNOT00000072241
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr11_-_46693565 | 0.02 |
ENSRNOT00000044900
|
Impg2
|
interphotoreceptor matrix proteoglycan 2 |
| chr7_+_126736732 | 0.02 |
ENSRNOT00000022012
|
Gtse1
|
G-2 and S-phase expressed 1 |
| chrX_+_74497262 | 0.02 |
ENSRNOT00000003899
|
Zcchc13
|
zinc finger CCHC-type containing 13 |
| chr4_+_110699557 | 0.02 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr3_+_117421604 | 0.02 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr7_-_116920507 | 0.02 |
ENSRNOT00000048363
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr2_+_205592182 | 0.02 |
ENSRNOT00000081050
|
Dennd2c
|
DENN domain containing 2C |
| chr20_-_6195463 | 0.02 |
ENSRNOT00000092474
|
Pxt1
|
peroxisomal, testis specific 1 |
| chr15_+_57290849 | 0.02 |
ENSRNOT00000014909
|
Cpb2
|
carboxypeptidase B2 |
| chr3_+_38280489 | 0.02 |
ENSRNOT00000011239
|
AABR07052130.1
|
|
| chr16_+_80729400 | 0.02 |
ENSRNOT00000036383
|
Tdrp
|
testis development related protein |
| chr15_+_28319136 | 0.02 |
ENSRNOT00000048723
|
Tppp2
|
tubulin polymerization-promoting protein family member 2 |
| chr6_+_109939345 | 0.02 |
ENSRNOT00000013560
|
Ift43
|
intraflagellar transport 43 |
| chr15_-_49467174 | 0.02 |
ENSRNOT00000019209
|
Adam7
|
ADAM metallopeptidase domain 7 |
| chr8_-_114766803 | 0.02 |
ENSRNOT00000047563
|
Col6a4
|
collagen, type VI, alpha 4 |
| chr10_-_63219309 | 0.02 |
ENSRNOT00000076357
|
Blmh
|
bleomycin hydrolase |
| chr13_-_35511020 | 0.02 |
ENSRNOT00000003413
|
Ralb
|
RAS like proto-oncogene B |
| chr4_+_31333970 | 0.02 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chr6_-_99204590 | 0.02 |
ENSRNOT00000041872
ENSRNOT00000007109 |
Esr2
|
estrogen receptor 2 |
| chr20_-_4390436 | 0.02 |
ENSRNOT00000000497
ENSRNOT00000077655 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr4_-_23718047 | 0.02 |
ENSRNOT00000011359
|
RGD1564345
|
similar to RIKEN cDNA 4921511H03 |
| chr2_+_243422811 | 0.02 |
ENSRNOT00000014694
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr1_+_198089446 | 0.02 |
ENSRNOT00000026146
|
Sgf29
|
SAGA complex associated factor 29 |
| chr8_+_117126692 | 0.02 |
ENSRNOT00000083046
ENSRNOT00000085609 |
Usp4
|
ubiquitin specific peptidase 4 |
| chr1_+_61095171 | 0.02 |
ENSRNOT00000042702
|
Vom1r19
|
vomeronasal 1 receptor 19 |
| chr1_-_216750844 | 0.02 |
ENSRNOT00000054862
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr10_+_53595854 | 0.02 |
ENSRNOT00000044116
|
Sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr12_-_21450204 | 0.02 |
ENSRNOT00000065866
|
LOC100910636
|
zinc finger CW-type PWWP domain protein 1-like |
| chr4_+_119842013 | 0.02 |
ENSRNOT00000057286
|
Hmces
|
5-hydroxymethylcytosine (hmC) binding, ES cell-specific |
| chr16_+_19973636 | 0.02 |
ENSRNOT00000024413
|
Mvb12a
|
multivesicular body subunit 12A |
| chr7_-_12922112 | 0.02 |
ENSRNOT00000010938
|
Madcam1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr10_-_110701137 | 0.02 |
ENSRNOT00000074193
|
Znf750
|
zinc finger protein 750 |
| chr1_-_82004538 | 0.02 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr6_-_99273033 | 0.02 |
ENSRNOT00000088808
|
Tex21
|
testis expressed 21 |
| chr20_-_32258286 | 0.02 |
ENSRNOT00000082114
|
Ddx50
|
DExD-box helicase 50 |
| chr5_+_173340060 | 0.02 |
ENSRNOT00000067841
|
Acap3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr4_-_119591817 | 0.02 |
ENSRNOT00000048768
|
AC097129.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.2 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:0060661 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.1 | GO:0002884 | negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.0 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.0 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.1 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |