Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
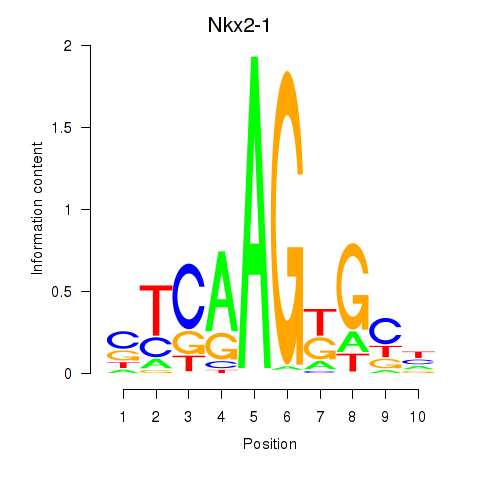
Results for Nkx2-1
Z-value: 0.47
Transcription factors associated with Nkx2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx2-1
|
ENSRNOG00000008644 | NK2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx2-1 | rn6_v1_chr6_-_77421286_77421286 | 0.86 | 6.3e-02 | Click! |
Activity profile of Nkx2-1 motif
Sorted Z-values of Nkx2-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_28654733 | 0.62 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr10_-_90307658 | 0.25 |
ENSRNOT00000092102
|
Slc4a1
|
solute carrier family 4 member 1 |
| chrX_+_136460215 | 0.19 |
ENSRNOT00000093538
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr1_+_279798187 | 0.15 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr10_+_40247436 | 0.14 |
ENSRNOT00000079830
|
Gpx3
|
glutathione peroxidase 3 |
| chr1_+_243589607 | 0.13 |
ENSRNOT00000021792
|
Dmrt3
|
doublesex and mab-3 related transcription factor 3 |
| chr17_-_43640387 | 0.12 |
ENSRNOT00000087731
|
Hist1h1c
|
histone cluster 1 H1 family member c |
| chr15_+_10120206 | 0.11 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr9_+_82647071 | 0.11 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr5_-_127273656 | 0.10 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr15_-_34198921 | 0.09 |
ENSRNOT00000024991
|
Nrl
|
neural retina leucine zipper |
| chr1_+_33910912 | 0.09 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr2_+_202200797 | 0.08 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr5_-_145423172 | 0.08 |
ENSRNOT00000081919
|
Gjb5
|
gap junction protein, beta 5 |
| chr1_+_86949413 | 0.08 |
ENSRNOT00000064153
|
Sirt2
|
sirtuin 2 |
| chr1_+_216237697 | 0.05 |
ENSRNOT00000027760
|
Cd81
|
Cd81 molecule |
| chr3_-_168018410 | 0.04 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr10_-_12990405 | 0.04 |
ENSRNOT00000078647
|
Thoc6
|
THO complex 6 |
| chr1_-_233140237 | 0.04 |
ENSRNOT00000083372
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr6_-_145777767 | 0.04 |
ENSRNOT00000046799
|
Rpl32
|
ribosomal protein L32 |
| chr12_+_11814071 | 0.04 |
ENSRNOT00000039149
|
Tmem130
|
transmembrane protein 130 |
| chr6_-_49851677 | 0.04 |
ENSRNOT00000007287
ENSRNOT00000077442 |
Acp1
|
acid phosphatase 1, soluble |
| chr6_-_136145837 | 0.04 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr10_-_84789832 | 0.03 |
ENSRNOT00000071719
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr13_-_82005741 | 0.03 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr12_+_25305823 | 0.03 |
ENSRNOT00000002010
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr7_+_141355994 | 0.02 |
ENSRNOT00000081195
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr10_-_65820538 | 0.02 |
ENSRNOT00000011808
|
Tmem97
|
transmembrane protein 97 |
| chr6_+_128973956 | 0.02 |
ENSRNOT00000075399
|
Fam181a
|
family with sequence similarity 181, member A |
| chr11_-_78456501 | 0.02 |
ENSRNOT00000036193
|
Tp63
|
tumor protein p63 |
| chr12_-_13462038 | 0.02 |
ENSRNOT00000043110
|
LOC100360439
|
ribosomal protein L36-like |
| chr7_-_55604403 | 0.02 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr1_+_170383682 | 0.02 |
ENSRNOT00000024224
|
Smpd1
|
sphingomyelin phosphodiesterase 1 |
| chr1_-_141470380 | 0.02 |
ENSRNOT00000065759
|
Plin1
|
perilipin 1 |
| chr1_-_227441442 | 0.02 |
ENSRNOT00000028433
|
Ms4a1
|
membrane spanning 4-domains A1 |
| chr7_+_116745061 | 0.02 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr5_-_22769907 | 0.02 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr11_-_78456200 | 0.02 |
ENSRNOT00000067251
ENSRNOT00000036179 |
Tp63
|
tumor protein p63 |
| chr7_-_11406771 | 0.02 |
ENSRNOT00000047450
|
Eef2
|
eukaryotic translation elongation factor 2 |
| chr13_+_52887649 | 0.01 |
ENSRNOT00000048033
|
Ascl5
|
achaete-scute family bHLH transcription factor 5 |
| chr15_-_51386190 | 0.01 |
ENSRNOT00000022706
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr7_+_107467260 | 0.01 |
ENSRNOT00000009240
ENSRNOT00000077382 |
Tg
|
thyroglobulin |
| chr2_-_250232295 | 0.01 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr12_+_41486076 | 0.01 |
ENSRNOT00000057242
|
Rita1
|
RBPJ interacting and tubulin associated 1 |
| chr6_+_101923785 | 0.01 |
ENSRNOT00000012005
|
Mpp5
|
membrane palmitoylated protein 5 |
| chr10_-_90096096 | 0.01 |
ENSRNOT00000081539
|
Tmem101
|
transmembrane protein 101 |
| chr9_+_10061978 | 0.01 |
ENSRNOT00000075180
|
Acer1
|
alkaline ceramidase 1 |
| chr9_+_81427730 | 0.01 |
ENSRNOT00000019109
ENSRNOT00000081711 |
Cxcr2
|
C-X-C motif chemokine receptor 2 |
| chr6_+_101924000 | 0.01 |
ENSRNOT00000092157
|
Mpp5
|
membrane palmitoylated protein 5 |
| chr10_+_89069256 | 0.01 |
ENSRNOT00000008576
|
Tubg2
|
tubulin, gamma 2 |
| chr2_+_188844073 | 0.01 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chr16_-_2227258 | 0.01 |
ENSRNOT00000089377
ENSRNOT00000067464 |
Slmap
|
sarcolemma associated protein |
| chrX_-_123600890 | 0.00 |
ENSRNOT00000067942
|
Sept6
|
septin 6 |
| chr4_-_70934295 | 0.00 |
ENSRNOT00000020616
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr4_+_100166863 | 0.00 |
ENSRNOT00000014505
|
Sftpb
|
surfactant protein B |
| chr1_-_197590277 | 0.00 |
ENSRNOT00000022502
ENSRNOT00000043433 |
Xpo6
|
exportin 6 |
| chr10_+_11240138 | 0.00 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx2-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.3 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:1904350 | regulation of protein catabolic process in the vacuole(GO:1904350) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |