Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
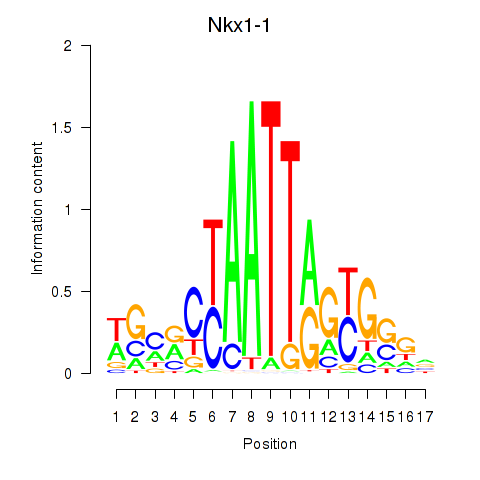
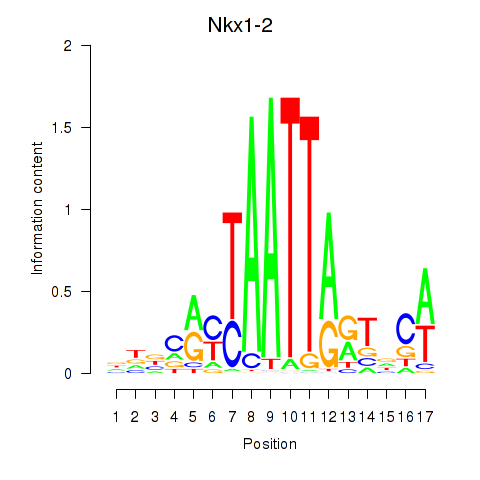
Results for Nkx1-1_Nkx1-2
Z-value: 0.28
Transcription factors associated with Nkx1-1_Nkx1-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nkx1-1
|
ENSRNOG00000005113 | NK1 homeobox 1 |
|
Nkx1-2
|
ENSRNOG00000017006 | NK1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nkx1-2 | rn6_v1_chr1_-_204605552_204605552 | 0.57 | 3.2e-01 | Click! |
Activity profile of Nkx1-1_Nkx1-2 motif
Sorted Z-values of Nkx1-1_Nkx1-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_35979193 | 0.20 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr2_+_145174876 | 0.19 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr5_-_168734296 | 0.16 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chrX_+_15113878 | 0.15 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr1_+_278557792 | 0.14 |
ENSRNOT00000023414
|
Atrnl1
|
attractin like 1 |
| chr10_+_103395511 | 0.11 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr3_+_14889510 | 0.09 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr9_+_10013854 | 0.08 |
ENSRNOT00000077653
ENSRNOT00000072033 |
Khsrp
|
KH-type splicing regulatory protein |
| chr19_+_45938915 | 0.07 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr2_-_33025271 | 0.07 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_+_2752680 | 0.06 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr4_+_88694583 | 0.06 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr3_-_120076788 | 0.06 |
ENSRNOT00000047158
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr9_+_71915421 | 0.05 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr11_-_51202703 | 0.05 |
ENSRNOT00000002719
ENSRNOT00000077220 |
Cblb
|
Cbl proto-oncogene B |
| chr9_+_73418607 | 0.05 |
ENSRNOT00000092547
|
Map2
|
microtubule-associated protein 2 |
| chr20_+_6211420 | 0.05 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr17_+_63635086 | 0.05 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr1_+_282134981 | 0.05 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr3_+_62648447 | 0.05 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr8_+_32188617 | 0.05 |
ENSRNOT00000081145
|
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr1_-_64147251 | 0.05 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr10_+_103206014 | 0.05 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr2_-_196177775 | 0.04 |
ENSRNOT00000028553
|
Zfp687
|
zinc finger protein 687 |
| chr1_-_215284548 | 0.04 |
ENSRNOT00000090375
|
Cttn
|
cortactin |
| chr2_-_188718704 | 0.04 |
ENSRNOT00000028010
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr12_+_49665282 | 0.04 |
ENSRNOT00000086397
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr1_+_141767940 | 0.04 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr17_-_22678253 | 0.04 |
ENSRNOT00000083122
|
LOC100362172
|
LRRGT00112-like |
| chr16_-_10802512 | 0.04 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr20_+_12944786 | 0.03 |
ENSRNOT00000048218
|
Pcnt
|
pericentrin |
| chr1_-_100926335 | 0.03 |
ENSRNOT00000020304
|
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr16_+_7035068 | 0.03 |
ENSRNOT00000024296
|
Nek4
|
NIMA-related kinase 4 |
| chr9_-_43022998 | 0.03 |
ENSRNOT00000063781
ENSRNOT00000089843 |
Lman2l
|
lectin, mannose-binding 2-like |
| chr7_-_12673659 | 0.03 |
ENSRNOT00000091650
ENSRNOT00000041277 ENSRNOT00000044865 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr1_+_15834779 | 0.03 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr16_+_7035241 | 0.03 |
ENSRNOT00000084977
|
Nek4
|
NIMA-related kinase 4 |
| chr5_+_131380297 | 0.03 |
ENSRNOT00000010548
|
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr6_+_111076351 | 0.03 |
ENSRNOT00000089644
|
Tmem63c
|
transmembrane protein 63c |
| chr17_-_87826421 | 0.03 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr2_+_148765412 | 0.03 |
ENSRNOT00000018248
ENSRNOT00000084662 |
Selenot
|
selenoprotein T |
| chr8_-_21492801 | 0.03 |
ENSRNOT00000077465
|
Zfp426
|
zinc finger protein 426 |
| chr1_-_198450047 | 0.03 |
ENSRNOT00000027360
|
Mvp
|
major vault protein |
| chr10_-_65502936 | 0.03 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr13_+_92569785 | 0.03 |
ENSRNOT00000082700
|
Fmn2
|
formin 2 |
| chr17_-_78812111 | 0.02 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr13_+_91054974 | 0.02 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr2_-_219458271 | 0.02 |
ENSRNOT00000064735
|
Cdc14a
|
cell division cycle 14A |
| chr9_+_3896337 | 0.02 |
ENSRNOT00000079166
|
LOC103693189
|
protein tyrosine phosphatase type IVA 1 |
| chr14_+_82375181 | 0.02 |
ENSRNOT00000023904
|
Fam53a
|
family with sequence similarity 53, member A |
| chrX_+_73390903 | 0.02 |
ENSRNOT00000077002
|
Zfp449
|
zinc finger protein 449 |
| chrX_+_140878216 | 0.02 |
ENSRNOT00000075840
ENSRNOT00000077026 |
Zic3
|
Zic family member 3 |
| chr7_-_91538673 | 0.02 |
ENSRNOT00000006209
|
Rad21
|
RAD21 cohesin complex component |
| chr13_-_83457888 | 0.02 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr1_-_101819478 | 0.02 |
ENSRNOT00000056181
|
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr4_+_158088505 | 0.02 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr1_-_134867001 | 0.02 |
ENSRNOT00000090149
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr5_+_71742911 | 0.02 |
ENSRNOT00000047225
|
Zfp462
|
zinc finger protein 462 |
| chr5_-_154222702 | 0.01 |
ENSRNOT00000080069
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr7_+_71157664 | 0.01 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr12_-_46493203 | 0.01 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr11_-_60882379 | 0.01 |
ENSRNOT00000002799
|
Cd200r1
|
CD200 receptor 1 |
| chr6_-_108660063 | 0.01 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr8_+_70789256 | 0.01 |
ENSRNOT00000048302
|
Clpx
|
caseinolytic mitochondrial matrix peptidase chaperone subunit |
| chr3_-_90751055 | 0.01 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr1_-_89007041 | 0.01 |
ENSRNOT00000032363
|
Proser3
|
proline and serine rich 3 |
| chr13_+_67611708 | 0.01 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr1_-_134871167 | 0.01 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr8_-_78096302 | 0.01 |
ENSRNOT00000086185
ENSRNOT00000077999 |
Myzap
|
myocardial zonula adherens protein |
| chr2_-_210088949 | 0.01 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr15_-_80713153 | 0.01 |
ENSRNOT00000063800
|
Klhl1
|
kelch-like family member 1 |
| chr1_+_261180007 | 0.01 |
ENSRNOT00000072181
|
Zdhhc16
|
zinc finger, DHHC-type containing 16 |
| chr12_-_12738620 | 0.01 |
ENSRNOT00000078603
ENSRNOT00000067021 |
Pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr6_+_18880737 | 0.01 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr20_-_13994794 | 0.01 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr3_+_80081647 | 0.01 |
ENSRNOT00000057161
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr10_+_48569555 | 0.01 |
ENSRNOT00000003966
|
Adora2b
|
adenosine A2B receptor |
| chr5_+_36566783 | 0.01 |
ENSRNOT00000077650
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr5_+_144114012 | 0.01 |
ENSRNOT00000035597
|
Stk40
|
serine/threonine kinase 40 |
| chr1_-_127292090 | 0.01 |
ENSRNOT00000016959
|
Lrrk1
|
leucine-rich repeat kinase 1 |
| chr16_+_39144972 | 0.01 |
ENSRNOT00000086728
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr14_-_7863664 | 0.00 |
ENSRNOT00000061162
|
Ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr5_+_58995249 | 0.00 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr2_-_149432106 | 0.00 |
ENSRNOT00000051027
|
P2ry13
|
purinergic receptor P2Y13 |
| chr7_+_117055395 | 0.00 |
ENSRNOT00000091960
|
Mapk15
|
mitogen-activated protein kinase 15 |
| chr6_-_44361908 | 0.00 |
ENSRNOT00000009491
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr1_-_80544825 | 0.00 |
ENSRNOT00000057802
ENSRNOT00000040060 ENSRNOT00000067049 ENSRNOT00000052387 ENSRNOT00000073352 |
Relb
|
RELB proto-oncogene, NF-kB subunit |
| chr4_+_14001761 | 0.00 |
ENSRNOT00000076519
|
Cd36
|
CD36 molecule |
| chr1_+_88113445 | 0.00 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr14_-_16903242 | 0.00 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr4_-_55011415 | 0.00 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr14_+_81858737 | 0.00 |
ENSRNOT00000029784
ENSRNOT00000058068 ENSRNOT00000058067 |
Poln
|
DNA polymerase nu |
| chr14_-_113644779 | 0.00 |
ENSRNOT00000005306
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr18_+_4084228 | 0.00 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr4_-_88684415 | 0.00 |
ENSRNOT00000009001
|
LOC500148
|
similar to 40S ribosomal protein S7 (S8) |
| chr2_+_248398917 | 0.00 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nkx1-1_Nkx1-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.0 | GO:0046154 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.0 | 0.0 | GO:0048382 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) mesendoderm development(GO:0048382) regulation of cardiac ventricle development(GO:1904412) |
| 0.0 | 0.0 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |