Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
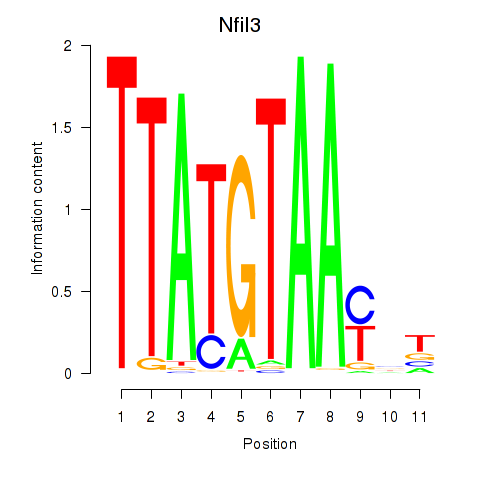
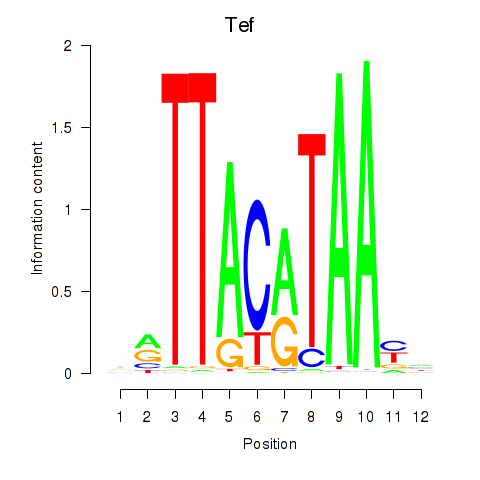
Results for Nfil3_Tef
Z-value: 1.01
Transcription factors associated with Nfil3_Tef
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfil3
|
ENSRNOG00000011668 | nuclear factor, interleukin 3 regulated |
|
Tef
|
ENSRNOG00000019383 | TEF, PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfil3 | rn6_v1_chr17_+_12261102_12261102 | -0.83 | 8.3e-02 | Click! |
| Tef | rn6_v1_chr7_+_123043503_123043584 | 0.65 | 2.4e-01 | Click! |
Activity profile of Nfil3_Tef motif
Sorted Z-values of Nfil3_Tef motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_168123395 | 0.54 |
ENSRNOT00000024932
|
Per3
|
period circadian clock 3 |
| chr18_-_26211445 | 0.44 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr6_+_43743702 | 0.43 |
ENSRNOT00000082570
|
Grhl1
|
grainyhead-like transcription factor 1 |
| chr3_+_95715193 | 0.43 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr6_-_105160470 | 0.41 |
ENSRNOT00000008367
|
Adam4
|
a disintegrin and metalloprotease domain 4 |
| chr10_-_16752205 | 0.40 |
ENSRNOT00000079462
|
Crebrf
|
CREB3 regulatory factor |
| chrX_-_10218583 | 0.40 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr15_-_47442664 | 0.37 |
ENSRNOT00000072994
|
Prss55
|
protease, serine, 55 |
| chr15_+_40665041 | 0.34 |
ENSRNOT00000018300
|
Amer2
|
APC membrane recruitment protein 2 |
| chr5_-_7941822 | 0.33 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr9_+_73529612 | 0.33 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr11_+_62584959 | 0.30 |
ENSRNOT00000071065
|
Gramd1c
|
GRAM domain containing 1C |
| chr8_-_36764422 | 0.29 |
ENSRNOT00000017330
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr10_+_65552897 | 0.25 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr15_+_8730871 | 0.25 |
ENSRNOT00000081845
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr4_-_68597586 | 0.24 |
ENSRNOT00000015921
|
RGD1563986
|
similar to RIKEN cDNA E330009J07 gene |
| chr3_+_111826297 | 0.23 |
ENSRNOT00000081897
|
Pla2g4b
|
phospholipase A2 group IVB |
| chr12_+_8725517 | 0.23 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chr9_-_44786041 | 0.22 |
ENSRNOT00000067197
|
Rev1
|
REV1, DNA directed polymerase |
| chr2_-_30634243 | 0.21 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr1_+_28454966 | 0.20 |
ENSRNOT00000078841
ENSRNOT00000030327 |
Tpd52l1
|
tumor protein D52-like 1 |
| chr1_+_198214797 | 0.20 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr3_+_114355798 | 0.20 |
ENSRNOT00000024658
ENSRNOT00000036435 |
Slc28a2
|
solute carrier family 28 member 2 |
| chr1_+_217345545 | 0.20 |
ENSRNOT00000071741
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr8_+_52753011 | 0.19 |
ENSRNOT00000047264
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr1_-_43638161 | 0.18 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr15_-_38165736 | 0.18 |
ENSRNOT00000063962
ENSRNOT00000014820 |
Zdhhc20
|
zinc finger, DHHC-type containing 20 |
| chr12_+_4737817 | 0.17 |
ENSRNOT00000035992
|
AABR07035107.1
|
|
| chr5_+_58661049 | 0.17 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr12_-_29743705 | 0.17 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr12_-_24537313 | 0.17 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr1_+_15642153 | 0.16 |
ENSRNOT00000079845
|
Map7
|
microtubule-associated protein 7 |
| chr13_+_44475970 | 0.16 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chrX_+_33884499 | 0.16 |
ENSRNOT00000090041
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr1_+_99505677 | 0.16 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr20_-_26852199 | 0.16 |
ENSRNOT00000078739
|
Sirt1
|
sirtuin 1 |
| chr3_-_146396299 | 0.16 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr1_+_15834779 | 0.16 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr10_-_5533695 | 0.16 |
ENSRNOT00000051564
|
Rpl39l
|
ribosomal protein L39-like |
| chr10_-_89700283 | 0.16 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr2_-_220838905 | 0.16 |
ENSRNOT00000078914
|
AABR07013065.1
|
|
| chr3_-_138683318 | 0.15 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr3_-_44086006 | 0.15 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr2_+_27005863 | 0.15 |
ENSRNOT00000080864
|
Poc5
|
POC5 centriolar protein |
| chr10_-_90030639 | 0.15 |
ENSRNOT00000090456
|
Mpp2
|
membrane palmitoylated protein 2 |
| chr18_+_79773608 | 0.15 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr7_-_130107437 | 0.15 |
ENSRNOT00000055865
|
Hdac10
|
histone deacetylase 10 |
| chr12_-_5490935 | 0.14 |
ENSRNOT00000050885
|
Zfp958
|
zinc finger protein 958 |
| chr13_-_98078555 | 0.14 |
ENSRNOT00000078034
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr1_-_217720414 | 0.14 |
ENSRNOT00000078810
|
Ppfia1
|
PTPRF interacting protein alpha 1 |
| chr10_-_89699836 | 0.14 |
ENSRNOT00000084311
|
Etv4
|
ets variant 4 |
| chr5_+_120340646 | 0.14 |
ENSRNOT00000086259
ENSRNOT00000086539 |
Dnajc6
|
DnaJ heat shock protein family (Hsp40) member C6 |
| chr6_+_101924000 | 0.14 |
ENSRNOT00000092157
|
Mpp5
|
membrane palmitoylated protein 5 |
| chrX_+_122938009 | 0.14 |
ENSRNOT00000089266
ENSRNOT00000017550 |
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr3_-_61488696 | 0.14 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr10_-_35187416 | 0.13 |
ENSRNOT00000084509
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr8_-_48597867 | 0.13 |
ENSRNOT00000077958
|
Nlrx1
|
NLR family member X1 |
| chr11_+_88732381 | 0.13 |
ENSRNOT00000078367
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr11_-_1836897 | 0.13 |
ENSRNOT00000050533
|
Zfp654
|
zinc finger protein 654 |
| chr2_+_252771017 | 0.13 |
ENSRNOT00000075452
ENSRNOT00000055318 |
Ttll7
|
tubulin tyrosine ligase like 7 |
| chr9_+_12475006 | 0.12 |
ENSRNOT00000079703
|
LOC100912293
|
uncharacterized LOC100912293 |
| chr2_-_43067956 | 0.12 |
ENSRNOT00000090616
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr13_-_78957183 | 0.12 |
ENSRNOT00000076954
ENSRNOT00000003851 |
Klhl20
|
kelch-like family member 20 |
| chrX_+_9436707 | 0.12 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr3_-_164964702 | 0.12 |
ENSRNOT00000014595
|
Adnp
|
activity-dependent neuroprotector homeobox |
| chr7_+_130308532 | 0.11 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr9_+_25410669 | 0.11 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr17_-_15566332 | 0.11 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chr11_+_17538063 | 0.11 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr7_-_59587382 | 0.11 |
ENSRNOT00000091427
ENSRNOT00000066396 |
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr2_-_195678848 | 0.11 |
ENSRNOT00000028303
ENSRNOT00000075569 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr13_-_98078946 | 0.11 |
ENSRNOT00000035875
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr12_-_35979193 | 0.10 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr15_+_34216833 | 0.10 |
ENSRNOT00000025260
|
Pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr3_+_58965552 | 0.10 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr5_+_104394119 | 0.10 |
ENSRNOT00000093557
ENSRNOT00000093462 |
Adamtsl1
|
ADAMTS-like 1 |
| chr14_-_14192048 | 0.10 |
ENSRNOT00000060868
|
Bmp2k
|
BMP2 inducible kinase |
| chr4_+_157107469 | 0.10 |
ENSRNOT00000015678
|
C1rl
|
complement C1r subcomponent like |
| chr4_+_147929863 | 0.10 |
ENSRNOT00000015427
|
LOC680385
|
similar to Sjogren syndrome antigen B |
| chr1_+_27476375 | 0.09 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr5_-_160742479 | 0.09 |
ENSRNOT00000019447
|
Kazn
|
kazrin, periplakin interacting protein |
| chr3_+_124515978 | 0.09 |
ENSRNOT00000028881
|
Prnp
|
prion protein |
| chr15_-_59215803 | 0.09 |
ENSRNOT00000032301
|
Lacc1
|
laccase domain containing 1 |
| chr8_+_116804451 | 0.09 |
ENSRNOT00000041402
|
Ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr13_-_84759098 | 0.09 |
ENSRNOT00000058683
|
LOC685351
|
similar to flavin containing monooxygenase 5 |
| chr14_-_62595854 | 0.09 |
ENSRNOT00000003024
|
Yipf7
|
Yip1 domain family, member 7 |
| chr14_+_66598259 | 0.09 |
ENSRNOT00000049743
|
Kcnip4
|
potassium voltage-gated channel interacting protein 4 |
| chr2_+_3662763 | 0.09 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr5_-_150163395 | 0.09 |
ENSRNOT00000079935
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr16_+_54332660 | 0.08 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr4_-_85915099 | 0.08 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr11_-_17119582 | 0.08 |
ENSRNOT00000087665
|
RGD1563888
|
similar to DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr13_+_74456487 | 0.08 |
ENSRNOT00000065801
|
Angptl1
|
angiopoietin-like 1 |
| chr14_+_79261092 | 0.08 |
ENSRNOT00000029191
|
LOC680039
|
hypothetical protein LOC680039 |
| chr13_-_111972603 | 0.08 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr1_-_204817080 | 0.08 |
ENSRNOT00000077956
|
Fam53b
|
family with sequence similarity 53, member B |
| chr12_+_18936594 | 0.08 |
ENSRNOT00000081587
|
Spry3
|
sprouty RTK signaling antagonist 3 |
| chr2_-_247988462 | 0.08 |
ENSRNOT00000022387
|
Pdlim5
|
PDZ and LIM domain 5 |
| chr8_-_39190457 | 0.08 |
ENSRNOT00000090161
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr7_-_59547174 | 0.08 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr10_-_56289882 | 0.07 |
ENSRNOT00000090762
ENSRNOT00000056903 |
Tnfsf13
|
tumor necrosis factor superfamily member 13 |
| chr15_+_105640097 | 0.07 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chr2_-_53300404 | 0.07 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr17_-_15484269 | 0.07 |
ENSRNOT00000020648
|
Omd
|
osteomodulin |
| chr13_-_82295123 | 0.07 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr16_+_65256751 | 0.07 |
ENSRNOT00000092012
|
AABR07026137.2
|
|
| chr9_-_9142339 | 0.07 |
ENSRNOT00000046852
|
MGC116197
|
similar to RIKEN cDNA 1700001E04 |
| chr2_-_178297172 | 0.07 |
ENSRNOT00000038543
|
Fnip2
|
folliculin interacting protein 2 |
| chr9_-_76768770 | 0.07 |
ENSRNOT00000087779
ENSRNOT00000057849 |
Ikzf2
|
IKAROS family zinc finger 2 |
| chr15_-_61648267 | 0.07 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr5_-_146787676 | 0.07 |
ENSRNOT00000008887
|
Zscan20
|
zinc finger and SCAN domain containing 20 |
| chr15_-_108376221 | 0.07 |
ENSRNOT00000034560
|
Gpr183
|
G protein-coupled receptor 183 |
| chr11_+_42259761 | 0.07 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr14_+_16924960 | 0.07 |
ENSRNOT00000003009
|
Ccdc158
|
coiled-coil domain containing 158 |
| chr1_+_61268248 | 0.07 |
ENSRNOT00000082730
|
LOC102552527
|
zinc finger protein 420-like |
| chr2_+_247248407 | 0.07 |
ENSRNOT00000082287
|
Unc5c
|
unc-5 netrin receptor C |
| chr2_-_18531210 | 0.07 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr12_+_504007 | 0.07 |
ENSRNOT00000001475
|
Brca2
|
BRCA2, DNA repair associated |
| chr19_+_15195565 | 0.07 |
ENSRNOT00000090865
ENSRNOT00000078874 |
Ces1d
|
carboxylesterase 1D |
| chrX_-_75224268 | 0.06 |
ENSRNOT00000089809
|
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr7_+_66742987 | 0.06 |
ENSRNOT00000045130
|
Mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr18_+_30496318 | 0.06 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr5_-_164502469 | 0.06 |
ENSRNOT00000051887
|
RGD1565622
|
RGD1565622 |
| chr9_+_117737611 | 0.06 |
ENSRNOT00000022396
|
Zfp161
|
zinc finger protein 161 |
| chr1_+_154579949 | 0.06 |
ENSRNOT00000045048
|
Sytl2
|
synaptotagmin-like 2 |
| chr9_+_73378057 | 0.06 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr16_-_45929 | 0.06 |
ENSRNOT00000043153
|
AABR07024439.1
|
|
| chr7_-_117364697 | 0.06 |
ENSRNOT00000077314
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr2_-_157066781 | 0.06 |
ENSRNOT00000001187
|
LOC304239
|
similar to RalA binding protein 1 |
| chr16_+_54291251 | 0.06 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr8_-_58195884 | 0.06 |
ENSRNOT00000010573
|
Acat1
|
acetyl-CoA acetyltransferase 1 |
| chr1_-_62558033 | 0.06 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr3_-_82777814 | 0.06 |
ENSRNOT00000068585
|
Accsl
|
1-aminocyclopropane-1-carboxylate synthase-like |
| chr3_-_46726946 | 0.06 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr1_+_30863217 | 0.06 |
ENSRNOT00000015421
|
Rnf146
|
ring finger protein 146 |
| chr15_+_31950986 | 0.06 |
ENSRNOT00000080233
|
AABR07017868.4
|
|
| chr17_-_45154355 | 0.06 |
ENSRNOT00000084976
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr15_+_24942218 | 0.06 |
ENSRNOT00000016629
|
Peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr15_+_55043608 | 0.06 |
ENSRNOT00000091000
|
Rcbtb2
|
RCC1 and BTB domain containing protein 2 |
| chr10_-_58693754 | 0.06 |
ENSRNOT00000071764
|
Pitpnm3
|
PITPNM family member 3 |
| chr16_+_66230841 | 0.05 |
ENSRNOT00000050464
|
LOC689479
|
similar to Discs large homolog 5 (Placenta and prostate DLG) (Discs large protein P-dlg) |
| chr8_-_94563760 | 0.05 |
ENSRNOT00000032792
|
Snap91
|
synaptosomal-associated protein 91 |
| chr3_+_177310753 | 0.05 |
ENSRNOT00000031448
|
Myt1
|
myelin transcription factor 1 |
| chr9_+_71915421 | 0.05 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr1_+_219759183 | 0.05 |
ENSRNOT00000026316
|
Pc
|
pyruvate carboxylase |
| chr6_-_137421283 | 0.05 |
ENSRNOT00000018596
|
Cdca4
|
cell division cycle associated 4 |
| chr10_+_94407559 | 0.05 |
ENSRNOT00000013046
|
Ddx42
|
DEAD-box helicase 42 |
| chr2_+_251200686 | 0.05 |
ENSRNOT00000019210
|
Col24a1
|
collagen type XXIV alpha 1 chain |
| chr1_+_94579080 | 0.05 |
ENSRNOT00000020079
|
LOC690000
|
similar to CG3740-PA |
| chr5_+_33580944 | 0.05 |
ENSRNOT00000092054
ENSRNOT00000036050 |
Rmdn1
|
regulator of microtubule dynamics 1 |
| chr10_-_55544429 | 0.05 |
ENSRNOT00000067528
|
Arhgef15
|
Rho guanine nucleotide exchange factor 15 |
| chr15_+_2766710 | 0.05 |
ENSRNOT00000017483
|
Dupd1
|
dual specificity phosphatase and pro isomerase domain containing 1 |
| chr14_+_48740190 | 0.05 |
ENSRNOT00000031638
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr10_-_90030423 | 0.05 |
ENSRNOT00000092150
|
Mpp2
|
membrane palmitoylated protein 2 |
| chr6_-_108660063 | 0.05 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chrX_-_31140711 | 0.05 |
ENSRNOT00000082358
|
Fancb
|
Fanconi anemia, complementation group B |
| chr12_-_8156151 | 0.05 |
ENSRNOT00000064721
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr17_-_76250537 | 0.05 |
ENSRNOT00000092693
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr5_-_151117042 | 0.05 |
ENSRNOT00000066549
|
Fam76a
|
family with sequence similarity 76, member A |
| chr5_-_113880911 | 0.04 |
ENSRNOT00000029441
|
Eqtn
|
equatorin |
| chr14_-_3300200 | 0.04 |
ENSRNOT00000037931
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr5_+_133819726 | 0.04 |
ENSRNOT00000081075
|
Stil
|
Scl/Tal1 interrupting locus |
| chr3_-_46051096 | 0.04 |
ENSRNOT00000081302
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr14_-_46153212 | 0.04 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr2_+_12516702 | 0.04 |
ENSRNOT00000050072
|
Tmem161b
|
transmembrane protein 161B |
| chr5_-_149987910 | 0.04 |
ENSRNOT00000091637
|
Ptpru
|
protein tyrosine phosphatase, receptor type, U |
| chrX_+_70645270 | 0.04 |
ENSRNOT00000076456
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr5_+_76386836 | 0.04 |
ENSRNOT00000088872
ENSRNOT00000021110 |
Ugcg
|
UDP-glucose ceramide glucosyltransferase |
| chr20_-_54517709 | 0.04 |
ENSRNOT00000076234
|
Grik2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr18_-_77317969 | 0.04 |
ENSRNOT00000090369
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr14_-_84751886 | 0.04 |
ENSRNOT00000078838
|
Mtmr3
|
myotubularin related protein 3 |
| chr1_+_57345577 | 0.04 |
ENSRNOT00000082790
ENSRNOT00000084017 |
Fam120b
|
family with sequence similarity 120B |
| chrX_-_138972684 | 0.04 |
ENSRNOT00000040165
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr9_-_19749145 | 0.04 |
ENSRNOT00000013956
|
Rcan2
|
regulator of calcineurin 2 |
| chr9_+_16749809 | 0.04 |
ENSRNOT00000028786
|
Cul9
|
cullin 9 |
| chr5_+_107323258 | 0.04 |
ENSRNOT00000008510
|
Klhl9
|
kelch-like family member 9 |
| chr19_-_10360310 | 0.04 |
ENSRNOT00000087100
|
Katnb1
|
katanin regulatory subunit B1 |
| chr3_+_158997834 | 0.04 |
ENSRNOT00000000211
|
LOC108348048
|
von Willebrand factor A domain-containing protein 5A |
| chr7_-_30274984 | 0.04 |
ENSRNOT00000092527
ENSRNOT00000010207 |
Slc17a8
|
solute carrier family 17 member 8 |
| chr6_+_86872903 | 0.04 |
ENSRNOT00000086894
|
Fancm
|
Fanconi anemia, complementation group M |
| chr3_+_124545364 | 0.04 |
ENSRNOT00000050900
|
Prnd
|
prion protein 2 (dublet) |
| chr9_-_91100482 | 0.04 |
ENSRNOT00000064642
|
LOC100911860
|
ubiquitin carboxyl-terminal hydrolase 40-like |
| chr4_-_168517177 | 0.04 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr5_-_50142724 | 0.04 |
ENSRNOT00000081150
|
Slc35a1
|
solute carrier family 35 member A1 |
| chr5_-_150525254 | 0.04 |
ENSRNOT00000075127
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr2_+_206454208 | 0.04 |
ENSRNOT00000026863
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr1_+_156262031 | 0.04 |
ENSRNOT00000038279
|
Tmem126b
|
transmembrane protein 126B |
| chr2_-_57935334 | 0.04 |
ENSRNOT00000022319
ENSRNOT00000085599 ENSRNOT00000077790 ENSRNOT00000035821 |
Slc1a3
|
solute carrier family 1 member 3 |
| chr1_+_255185629 | 0.04 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr3_+_79729739 | 0.04 |
ENSRNOT00000084833
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr6_+_73358112 | 0.03 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr5_+_151181559 | 0.03 |
ENSRNOT00000085674
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr1_+_217345154 | 0.03 |
ENSRNOT00000092516
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr14_+_19318565 | 0.03 |
ENSRNOT00000068682
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr2_-_210454737 | 0.03 |
ENSRNOT00000079993
|
Ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr8_-_17696989 | 0.03 |
ENSRNOT00000083752
|
AABR07069336.1
|
|
| chr13_-_51784639 | 0.03 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr19_+_15033108 | 0.03 |
ENSRNOT00000021812
|
Ces1d
|
carboxylesterase 1D |
| chr2_-_244335165 | 0.03 |
ENSRNOT00000084860
|
Rap1gds1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr2_+_104020955 | 0.03 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfil3_Tef
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904937 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.1 | 0.4 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.1 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.2 | GO:2000612 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) maintenance of chromatin silencing(GO:0006344) thyroid-stimulating hormone secretion(GO:0070460) response to resveratrol(GO:1904638) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.3 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.4 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.2 | GO:0099525 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0061470 | astrocyte chemotaxis(GO:0035700) T follicular helper cell differentiation(GO:0061470) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0046952 | acetyl-CoA catabolic process(GO:0046356) ketone body catabolic process(GO:0046952) metanephric proximal tubule development(GO:0072237) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.0 | 0.2 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.5 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) negative regulation of long-term synaptic potentiation(GO:1900272) negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.0 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.0 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.3 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0098835 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.2 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0015489 | ornithine decarboxylase inhibitor activity(GO:0008073) polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.1 | GO:0052724 | inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.2 | GO:0098919 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |