Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
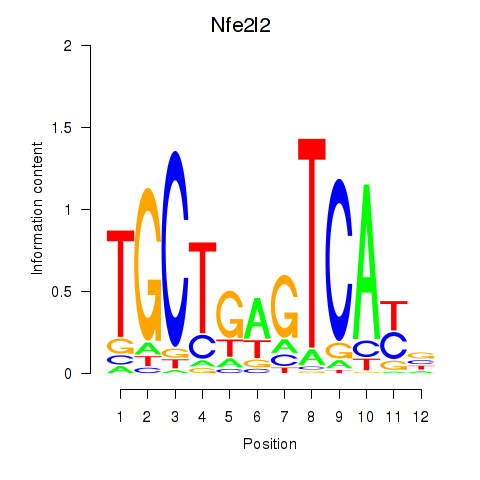
Results for Nfe2l2
Z-value: 0.54
Transcription factors associated with Nfe2l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2l2
|
ENSRNOG00000001548 | nuclear factor, erythroid 2-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2l2 | rn6_v1_chr3_-_62524996_62524996 | -0.38 | 5.3e-01 | Click! |
Activity profile of Nfe2l2 motif
Sorted Z-values of Nfe2l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_166943592 | 0.50 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr1_-_88881460 | 0.25 |
ENSRNOT00000028287
|
Hcst
|
hematopoietic cell signal transducer |
| chr11_+_87435185 | 0.22 |
ENSRNOT00000002558
|
P2rx6
|
purinergic receptor P2X 6 |
| chr2_+_115362799 | 0.18 |
ENSRNOT00000015756
|
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr4_-_44136815 | 0.17 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr14_-_115052450 | 0.17 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr13_-_90602365 | 0.16 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr2_+_24546536 | 0.16 |
ENSRNOT00000014036
|
Otp
|
orthopedia homeobox |
| chr11_+_45304068 | 0.15 |
ENSRNOT00000031464
|
Cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr3_-_2490392 | 0.14 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr2_+_225827504 | 0.13 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr8_+_76426335 | 0.13 |
ENSRNOT00000085496
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr6_+_128738388 | 0.13 |
ENSRNOT00000050204
|
Rpl6-ps1
|
ribosomal protein L6, pseudogene 1 |
| chr3_+_103747654 | 0.13 |
ENSRNOT00000006887
|
Nop10
|
NOP10 ribonucleoprotein |
| chr17_-_53102342 | 0.13 |
ENSRNOT00000073834
|
Psma2
|
proteasome subunit alpha 2 |
| chr2_-_196415530 | 0.13 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr10_-_61576952 | 0.12 |
ENSRNOT00000092712
|
Pafah1b1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 |
| chr10_+_84309430 | 0.11 |
ENSRNOT00000030159
|
Skap1
|
src kinase associated phosphoprotein 1 |
| chr1_-_219294147 | 0.11 |
ENSRNOT00000024601
|
Gstp1
|
glutathione S-transferase pi 1 |
| chr11_+_30363280 | 0.10 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr1_-_88123643 | 0.10 |
ENSRNOT00000027937
|
Psmd8
|
proteasome 26S subunit, non-ATPase 8 |
| chr15_-_33263659 | 0.10 |
ENSRNOT00000018005
|
Psmb5
|
proteasome subunit beta 5 |
| chr17_-_14687408 | 0.10 |
ENSRNOT00000088542
|
AC120310.1
|
|
| chr16_+_23317953 | 0.10 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr13_-_36101411 | 0.09 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr5_+_135536413 | 0.09 |
ENSRNOT00000023132
|
Prdx1
|
peroxiredoxin 1 |
| chr5_+_57947716 | 0.09 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chr3_-_10371240 | 0.09 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr7_+_119626637 | 0.09 |
ENSRNOT00000000201
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr3_-_175426395 | 0.09 |
ENSRNOT00000082686
|
Psma7
|
proteasome subunit alpha 7 |
| chr6_-_42302254 | 0.09 |
ENSRNOT00000007071
|
Pqlc3
|
PQ loop repeat containing 3 |
| chr20_+_295250 | 0.09 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chr7_+_79138925 | 0.08 |
ENSRNOT00000047511
|
AABR07057586.1
|
|
| chr6_+_93423002 | 0.08 |
ENSRNOT00000010753
|
Psma3
|
proteasome subunit alpha 3 |
| chr15_+_4351292 | 0.08 |
ENSRNOT00000009207
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chrX_+_159505344 | 0.08 |
ENSRNOT00000001164
|
Brs3
|
bombesin receptor subtype 3 |
| chr7_+_59200918 | 0.08 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr19_+_37282018 | 0.08 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chrX_+_28593405 | 0.08 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr8_+_133029625 | 0.07 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr9_+_17817721 | 0.07 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr4_+_3043231 | 0.07 |
ENSRNOT00000013732
|
Il6
|
interleukin 6 |
| chr4_+_66670618 | 0.07 |
ENSRNOT00000010796
|
Tbxas1
|
thromboxane A synthase 1 |
| chr6_+_76188083 | 0.07 |
ENSRNOT00000009666
|
Psma6
|
proteasome subunit alpha 6 |
| chr4_+_176994129 | 0.07 |
ENSRNOT00000018734
|
Cmas
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr11_+_53081025 | 0.07 |
ENSRNOT00000002700
|
Bbx
|
BBX, HMG-box containing |
| chr11_-_76888178 | 0.07 |
ENSRNOT00000049841
|
Ostn
|
osteocrin |
| chr1_-_170318935 | 0.07 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr18_-_1216379 | 0.07 |
ENSRNOT00000020537
ENSRNOT00000084613 |
Usp14
|
ubiquitin specific peptidase 14 |
| chr8_-_13290773 | 0.07 |
ENSRNOT00000012217
|
RGD1561795
|
similar to RIKEN cDNA 1700012B09 |
| chr1_-_57489727 | 0.07 |
ENSRNOT00000002037
|
Psmb1
|
proteasome subunit beta 1 |
| chr3_+_2490518 | 0.06 |
ENSRNOT00000015099
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr1_+_87066289 | 0.06 |
ENSRNOT00000027645
|
Capn12
|
calpain 12 |
| chr7_+_2479311 | 0.06 |
ENSRNOT00000003787
|
Ptges3
|
prostaglandin E synthase 3 |
| chr17_+_53102534 | 0.06 |
ENSRNOT00000021433
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr1_+_101324652 | 0.06 |
ENSRNOT00000028116
|
Hrc
|
histidine rich calcium binding protein |
| chr9_+_15166118 | 0.06 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr2_+_211050360 | 0.06 |
ENSRNOT00000026928
|
Psma5
|
proteasome subunit alpha 5 |
| chr4_-_28953067 | 0.06 |
ENSRNOT00000013989
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr10_+_95706685 | 0.05 |
ENSRNOT00000004283
|
Psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr5_+_144701949 | 0.05 |
ENSRNOT00000015618
|
Psmb2
|
proteasome subunit beta 2 |
| chr6_-_128690741 | 0.05 |
ENSRNOT00000035826
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr1_-_183744586 | 0.05 |
ENSRNOT00000015946
|
Psma1
|
proteasome subunit alpha 1 |
| chr11_-_83944763 | 0.05 |
ENSRNOT00000002358
|
Psmd2
|
proteasome 26S subunit, non-ATPase 2 |
| chr6_-_26051229 | 0.05 |
ENSRNOT00000005855
|
Bre
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr19_+_19395655 | 0.05 |
ENSRNOT00000019130
|
Snx20
|
sorting nexin 20 |
| chr14_+_22091777 | 0.05 |
ENSRNOT00000072483
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr2_+_46980976 | 0.05 |
ENSRNOT00000083668
ENSRNOT00000082990 |
Mocs2
|
molybdenum cofactor synthesis 2 |
| chr19_-_43848937 | 0.05 |
ENSRNOT00000044844
|
Ldhd
|
lactate dehydrogenase D |
| chr18_-_59086251 | 0.05 |
ENSRNOT00000025510
ENSRNOT00000083831 |
Txnl1
|
thioredoxin-like 1 |
| chr15_+_2526368 | 0.05 |
ENSRNOT00000048713
ENSRNOT00000074803 |
Dusp13
Dusp13
|
dual specificity phosphatase 13 dual specificity phosphatase 13 |
| chr3_-_159802952 | 0.05 |
ENSRNOT00000011610
|
Oser1
|
oxidative stress responsive serine-rich 1 |
| chr1_-_225128740 | 0.05 |
ENSRNOT00000026897
|
Rom1
|
retinal outer segment membrane protein 1 |
| chr13_+_60545635 | 0.04 |
ENSRNOT00000077153
|
Uchl5
|
ubiquitin C-terminal hydrolase L5 |
| chr8_-_115179191 | 0.04 |
ENSRNOT00000017224
|
Parp3
|
poly (ADP-ribose) polymerase family, member 3 |
| chr1_-_198232344 | 0.04 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr17_-_70481750 | 0.04 |
ENSRNOT00000025252
|
Il15ra
|
interleukin 15 receptor subunit alpha |
| chr15_-_30174197 | 0.04 |
ENSRNOT00000074419
|
LOC688340
|
T-cell receptor alpha chain V region PHDS58-like |
| chr11_-_84037938 | 0.04 |
ENSRNOT00000002327
|
Abcf3
|
ATP binding cassette subfamily F member 3 |
| chr7_-_120919261 | 0.04 |
ENSRNOT00000019612
|
Josd1
|
Josephin domain containing 1 |
| chr7_+_2737765 | 0.04 |
ENSRNOT00000004895
|
Cnpy2
|
canopy FGF signaling regulator 2 |
| chr7_+_64768742 | 0.04 |
ENSRNOT00000005545
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr3_-_5483239 | 0.04 |
ENSRNOT00000071754
|
Surf4
|
surfeit 4 |
| chr4_+_157511642 | 0.04 |
ENSRNOT00000065846
|
Pianp
|
PILR alpha associated neural protein |
| chr19_-_39646693 | 0.04 |
ENSRNOT00000019104
|
Psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr15_-_35113678 | 0.04 |
ENSRNOT00000059677
|
Ctsg
|
cathepsin G |
| chr8_+_33514042 | 0.04 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr2_-_187854363 | 0.04 |
ENSRNOT00000092993
|
Lmna
|
lamin A/C |
| chr10_+_46906115 | 0.03 |
ENSRNOT00000034006
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr13_+_27889345 | 0.03 |
ENSRNOT00000079134
ENSRNOT00000003274 |
Serpinb8
|
serpin family B member 8 |
| chr15_+_31141142 | 0.03 |
ENSRNOT00000060304
|
LOC103693854
|
uncharacterized LOC103693854 |
| chr8_+_80965255 | 0.03 |
ENSRNOT00000079508
|
Wdr72
|
WD repeat domain 72 |
| chr3_+_79876938 | 0.03 |
ENSRNOT00000015757
|
Psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr1_-_222734184 | 0.03 |
ENSRNOT00000049812
ENSRNOT00000028785 |
Rtn3
|
reticulon 3 |
| chrX_-_31968152 | 0.03 |
ENSRNOT00000004884
|
Pir
|
pirin |
| chr11_-_86328469 | 0.03 |
ENSRNOT00000071493
|
Ufd1l
|
ubiquitin fusion degradation 1 like (yeast) |
| chr13_+_92264231 | 0.03 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr6_-_99843245 | 0.03 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chrX_-_72034099 | 0.03 |
ENSRNOT00000004310
|
Ercc6l
|
ERCC excision repair 6 like, spindle assembly checkpoint helicase |
| chr10_-_63240332 | 0.03 |
ENSRNOT00000005125
|
Blmh
|
bleomycin hydrolase |
| chr17_-_66511870 | 0.03 |
ENSRNOT00000044662
|
Lgals8
|
galectin 8 |
| chr18_+_55666027 | 0.03 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr1_+_12915734 | 0.03 |
ENSRNOT00000089066
|
Txlnb
|
taxilin beta |
| chr8_-_130550388 | 0.03 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr3_+_146365092 | 0.03 |
ENSRNOT00000008865
|
Cst7
|
cystatin F |
| chr2_-_190100276 | 0.03 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr19_+_52077501 | 0.02 |
ENSRNOT00000079240
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr2_-_238246931 | 0.02 |
ENSRNOT00000086303
|
Gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr6_-_49089855 | 0.02 |
ENSRNOT00000006526
|
Tpo
|
thyroid peroxidase |
| chr1_+_85337805 | 0.02 |
ENSRNOT00000026839
|
Samd4b
|
sterile alpha motif domain containing 4B |
| chr11_+_86797557 | 0.02 |
ENSRNOT00000083049
ENSRNOT00000046594 |
Tango2
|
transport and golgi organization 2 homolog |
| chrX_+_17171605 | 0.02 |
ENSRNOT00000048236
|
Nudt10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr7_+_10937599 | 0.02 |
ENSRNOT00000008758
|
Sirt6
|
sirtuin 6 |
| chr16_-_14382641 | 0.02 |
ENSRNOT00000018723
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr11_-_81660395 | 0.02 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr5_-_101006375 | 0.02 |
ENSRNOT00000014008
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr4_+_162392869 | 0.02 |
ENSRNOT00000047790
|
Clec2d
|
C-type lectin domain family 2, member D |
| chr18_-_76984203 | 0.02 |
ENSRNOT00000089122
|
Ctdp1
|
CTD phosphatase subunit 1 |
| chr4_+_163293724 | 0.02 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chr10_+_46018659 | 0.02 |
ENSRNOT00000040865
ENSRNOT00000004357 |
Mprip
|
myosin phosphatase Rho interacting protein |
| chr20_+_5527181 | 0.02 |
ENSRNOT00000091364
|
Phf1
|
PHD finger protein 1 |
| chr10_+_94445877 | 0.02 |
ENSRNOT00000013997
|
Psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr20_-_41209728 | 0.02 |
ENSRNOT00000000657
|
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr7_-_121058029 | 0.02 |
ENSRNOT00000068033
|
Cbx6
|
chromobox 6 |
| chr9_-_13513960 | 0.02 |
ENSRNOT00000067626
|
Mocs1
|
molybdenum cofactor synthesis 1 |
| chr3_+_152294656 | 0.02 |
ENSRNOT00000027067
|
Phf20
|
PHD finger protein 20 |
| chr5_-_145418992 | 0.02 |
ENSRNOT00000037128
|
Gjb4
|
gap junction protein, beta 4 |
| chr15_+_44411865 | 0.02 |
ENSRNOT00000017566
|
Kctd9
|
potassium channel tetramerization domain containing 9 |
| chr5_-_167244786 | 0.02 |
ENSRNOT00000051309
|
Car6
|
carbonic anhydrase 6 |
| chr3_+_160231914 | 0.01 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr8_-_28075514 | 0.01 |
ENSRNOT00000072851
|
Vps26b
|
VPS26 retromer complex component B |
| chr13_-_85443727 | 0.01 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr17_-_9791781 | 0.01 |
ENSRNOT00000090536
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr5_-_58445953 | 0.01 |
ENSRNOT00000046102
|
Vcp
|
valosin-containing protein |
| chr19_+_38422164 | 0.01 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr4_+_7314369 | 0.01 |
ENSRNOT00000029724
|
Atg9b
|
autophagy related 9B |
| chr15_+_32832035 | 0.01 |
ENSRNOT00000060250
|
AABR07017902.1
|
|
| chr19_+_45946236 | 0.01 |
ENSRNOT00000045250
|
Syce1l
|
synaptonemal complex central element protein 1-like |
| chr19_+_52077109 | 0.01 |
ENSRNOT00000020225
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr16_-_62239987 | 0.01 |
ENSRNOT00000020252
|
Gsr
|
glutathione-disulfide reductase |
| chrX_+_104734082 | 0.01 |
ENSRNOT00000005020
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr9_+_51298426 | 0.01 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr5_-_150994913 | 0.01 |
ENSRNOT00000075506
|
RGD1561465
|
similar to RIKEN cDNA 2900010J23 |
| chr8_+_45797315 | 0.01 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr9_-_99659425 | 0.01 |
ENSRNOT00000051686
|
Olr1343
|
olfactory receptor 1343 |
| chr8_+_119228612 | 0.01 |
ENSRNOT00000078439
ENSRNOT00000043737 |
Lrrc2
|
leucine rich repeat containing 2 |
| chr1_-_165997751 | 0.01 |
ENSRNOT00000050227
|
P2ry6
|
pyrimidinergic receptor P2Y6 |
| chr17_-_9792007 | 0.01 |
ENSRNOT00000021596
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr5_+_64294321 | 0.01 |
ENSRNOT00000083796
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr10_+_105499569 | 0.01 |
ENSRNOT00000088457
|
Sphk1
|
sphingosine kinase 1 |
| chr1_+_199248470 | 0.01 |
ENSRNOT00000025933
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr12_-_52676608 | 0.01 |
ENSRNOT00000072507
|
Plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr3_-_2456041 | 0.00 |
ENSRNOT00000062001
|
Rnf224
|
ring finger protein 224 |
| chr20_+_3987230 | 0.00 |
ENSRNOT00000088615
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr1_+_199360645 | 0.00 |
ENSRNOT00000026527
|
Kat8
|
lysine acetyltransferase 8 |
| chr3_+_2648885 | 0.00 |
ENSRNOT00000020339
|
Abca2
|
ATP binding cassette subfamily A member 2 |
| chr19_-_37281933 | 0.00 |
ENSRNOT00000059663
|
Lrrc29
|
leucine rich repeat containing 29 |
| chr20_+_5526975 | 0.00 |
ENSRNOT00000059548
|
Phf1
|
PHD finger protein 1 |
| chr16_-_20383337 | 0.00 |
ENSRNOT00000025977
|
Il12rb1
|
interleukin 12 receptor subunit beta 1 |
| chrX_-_142248369 | 0.00 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr15_-_4035064 | 0.00 |
ENSRNOT00000012300
|
Fut11
|
fucosyltransferase 11 |
| chr3_-_122920684 | 0.00 |
ENSRNOT00000028818
|
Cpxm1
|
carboxypeptidase X (M14 family), member 1 |
| chr8_-_23282989 | 0.00 |
ENSRNOT00000019204
|
Zfp717
|
zinc finger protein 717 |
| chr1_+_199555722 | 0.00 |
ENSRNOT00000054983
|
Itgax
|
integrin subunit alpha X |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.1 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 0.0 | 0.1 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of chemokine biosynthetic process(GO:0045079) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.1 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.0 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.0 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.0 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.0 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) negative regulation of interleukin-8 secretion(GO:2000483) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0017098 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |