Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
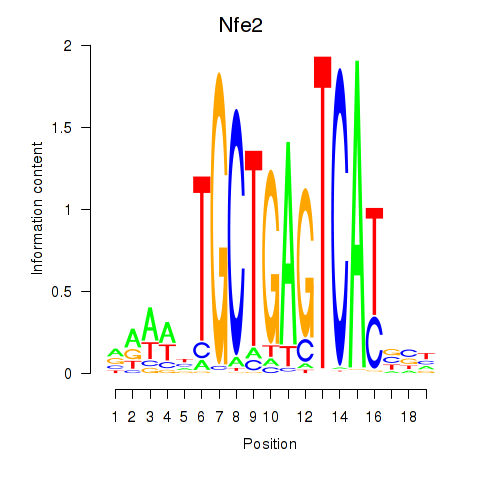
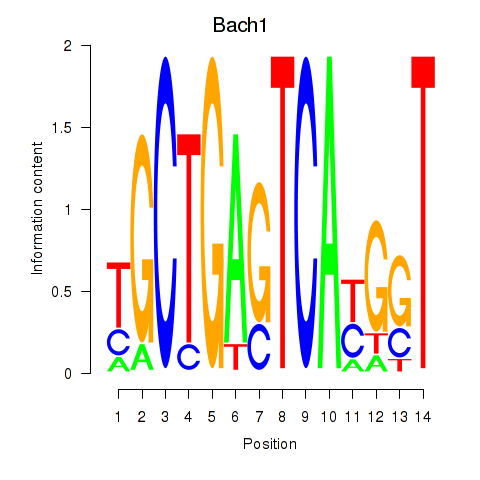
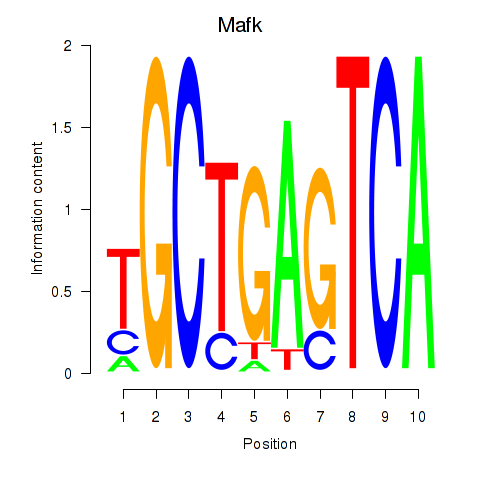
Results for Nfe2_Bach1_Mafk
Z-value: 0.62
Transcription factors associated with Nfe2_Bach1_Mafk
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2
|
ENSRNOG00000036837 | nuclear factor, erythroid 2 |
|
Bach1
|
ENSRNOG00000001582 | BTB domain and CNC homolog 1 |
|
Mafk
|
ENSRNOG00000001277 | MAF bZIP transcription factor K |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mafk | rn6_v1_chr12_-_16934706_16934706 | 0.50 | 3.9e-01 | Click! |
| Bach1 | rn6_v1_chr11_+_27364916_27364916 | 0.44 | 4.6e-01 | Click! |
| Nfe2 | rn6_v1_chr7_-_144880092_144880092 | -0.13 | 8.3e-01 | Click! |
Activity profile of Nfe2_Bach1_Mafk motif
Sorted Z-values of Nfe2_Bach1_Mafk motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_88551056 | 0.65 |
ENSRNOT00000032737
|
Zfp385c
|
zinc finger protein 385C |
| chr12_-_2174131 | 0.45 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr1_-_166943592 | 0.43 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr7_+_59200918 | 0.34 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr11_+_87435185 | 0.32 |
ENSRNOT00000002558
|
P2rx6
|
purinergic receptor P2X 6 |
| chr19_-_37250919 | 0.25 |
ENSRNOT00000021015
|
Exoc3l1
|
exocyst complex component 3-like 1 |
| chr1_-_88881460 | 0.22 |
ENSRNOT00000028287
|
Hcst
|
hematopoietic cell signal transducer |
| chr1_-_221917901 | 0.22 |
ENSRNOT00000092270
|
Slc22a12
|
solute carrier family 22 member 12 |
| chr1_+_198690794 | 0.20 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr17_-_53102342 | 0.17 |
ENSRNOT00000073834
|
Psma2
|
proteasome subunit alpha 2 |
| chr8_+_76426335 | 0.17 |
ENSRNOT00000085496
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr2_+_115362799 | 0.16 |
ENSRNOT00000015756
|
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr14_-_115052450 | 0.16 |
ENSRNOT00000067998
|
Acyp2
|
acylphosphatase 2 |
| chr3_-_10371240 | 0.16 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr12_+_50407843 | 0.15 |
ENSRNOT00000073763
|
Cryba4
|
crystallin, beta A4 |
| chr2_+_225827504 | 0.15 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr11_-_84037938 | 0.14 |
ENSRNOT00000002327
|
Abcf3
|
ATP binding cassette subfamily F member 3 |
| chr7_-_130120579 | 0.14 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr9_+_51298426 | 0.13 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr15_+_44411865 | 0.13 |
ENSRNOT00000017566
|
Kctd9
|
potassium channel tetramerization domain containing 9 |
| chr20_+_5008508 | 0.13 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr9_+_10963723 | 0.12 |
ENSRNOT00000075424
|
Plin4
|
perilipin 4 |
| chr13_-_36101411 | 0.12 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr7_+_125576327 | 0.12 |
ENSRNOT00000016679
|
Prr5
|
proline rich 5 |
| chr3_+_147609095 | 0.12 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr2_-_199479309 | 0.11 |
ENSRNOT00000042661
|
Rpl38
|
ribosomal protein L38 |
| chr6_-_99843245 | 0.11 |
ENSRNOT00000080270
|
Gpx2
|
glutathione peroxidase 2 |
| chr1_-_198232344 | 0.11 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr4_-_44136815 | 0.10 |
ENSRNOT00000086810
|
Tfec
|
transcription factor EC |
| chr11_+_30363280 | 0.09 |
ENSRNOT00000002885
|
Sod1
|
superoxide dismutase 1, soluble |
| chr11_-_86328469 | 0.09 |
ENSRNOT00000071493
|
Ufd1l
|
ubiquitin fusion degradation 1 like (yeast) |
| chr12_-_50404550 | 0.09 |
ENSRNOT00000073072
|
Crybb1
|
crystallin, beta B1 |
| chr8_+_48571323 | 0.09 |
ENSRNOT00000059776
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_-_219294147 | 0.09 |
ENSRNOT00000024601
|
Gstp1
|
glutathione S-transferase pi 1 |
| chr8_+_133029625 | 0.08 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr1_+_264507985 | 0.08 |
ENSRNOT00000085811
|
Pax2
|
paired box 2 |
| chr1_+_193187972 | 0.08 |
ENSRNOT00000090172
ENSRNOT00000065342 |
Slc5a11
|
solute carrier family 5 member 11 |
| chr9_-_82154266 | 0.08 |
ENSRNOT00000024283
|
Cryba2
|
crystallin, beta A2 |
| chr7_+_3355116 | 0.07 |
ENSRNOT00000030453
ENSRNOT00000061657 |
Itga7
|
integrin subunit alpha 7 |
| chr1_+_276659542 | 0.07 |
ENSRNOT00000019681
|
Tcf7l2
|
transcription factor 7 like 2 |
| chr14_+_34354503 | 0.07 |
ENSRNOT00000002941
|
Nmu
|
neuromedin U |
| chr5_+_135536413 | 0.07 |
ENSRNOT00000023132
|
Prdx1
|
peroxiredoxin 1 |
| chr10_-_61576952 | 0.07 |
ENSRNOT00000092712
|
Pafah1b1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 |
| chr6_-_42302254 | 0.06 |
ENSRNOT00000007071
|
Pqlc3
|
PQ loop repeat containing 3 |
| chr1_-_259357056 | 0.06 |
ENSRNOT00000022000
|
Pdlim1
|
PDZ and LIM domain 1 |
| chr5_-_150994913 | 0.06 |
ENSRNOT00000075506
|
RGD1561465
|
similar to RIKEN cDNA 2900010J23 |
| chr4_-_97784842 | 0.06 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr3_+_2648885 | 0.06 |
ENSRNOT00000020339
|
Abca2
|
ATP binding cassette subfamily A member 2 |
| chr15_-_44860604 | 0.06 |
ENSRNOT00000018637
|
Nefm
|
neurofilament medium |
| chr9_+_15166118 | 0.06 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr15_-_95514259 | 0.06 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr13_+_52854031 | 0.06 |
ENSRNOT00000013738
ENSRNOT00000086004 |
Tmem9
|
transmembrane protein 9 |
| chrX_+_156854594 | 0.06 |
ENSRNOT00000083442
|
Renbp
|
renin binding protein |
| chr10_-_35716260 | 0.06 |
ENSRNOT00000004308
ENSRNOT00000059255 |
Sqstm1
|
sequestosome 1 |
| chr20_-_3818045 | 0.06 |
ENSRNOT00000091622
|
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr15_+_33121273 | 0.06 |
ENSRNOT00000016020
|
Rem2
|
RRAD and GEM like GTPase 2 |
| chr19_+_52077109 | 0.06 |
ENSRNOT00000020225
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr6_+_128738388 | 0.06 |
ENSRNOT00000050204
|
Rpl6-ps1
|
ribosomal protein L6, pseudogene 1 |
| chr10_-_63240332 | 0.05 |
ENSRNOT00000005125
|
Blmh
|
bleomycin hydrolase |
| chr20_-_5479511 | 0.05 |
ENSRNOT00000000558
|
Zbtb22
|
zinc finger and BTB domain containing 22 |
| chr4_-_100099517 | 0.05 |
ENSRNOT00000014277
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr6_+_93423002 | 0.05 |
ENSRNOT00000010753
|
Psma3
|
proteasome subunit alpha 3 |
| chr19_+_57556694 | 0.05 |
ENSRNOT00000032359
|
Trim67
|
tripartite motif-containing 67 |
| chr3_-_2490392 | 0.05 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr10_-_18574710 | 0.05 |
ENSRNOT00000079031
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr17_-_66511870 | 0.05 |
ENSRNOT00000044662
|
Lgals8
|
galectin 8 |
| chr14_+_107750162 | 0.05 |
ENSRNOT00000012767
|
Zrsr1
|
zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 |
| chr18_+_30826260 | 0.05 |
ENSRNOT00000065235
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1 |
| chr17_-_70481750 | 0.05 |
ENSRNOT00000025252
|
Il15ra
|
interleukin 15 receptor subunit alpha |
| chr2_+_93792601 | 0.04 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr1_+_204959174 | 0.04 |
ENSRNOT00000023257
|
Zranb1
|
zinc finger RANBP2-type containing 1 |
| chr2_-_187854363 | 0.04 |
ENSRNOT00000092993
|
Lmna
|
lamin A/C |
| chr17_-_14687408 | 0.04 |
ENSRNOT00000088542
|
AC120310.1
|
|
| chr5_-_38923095 | 0.04 |
ENSRNOT00000009146
|
AABR07047593.1
|
|
| chr3_-_176958880 | 0.04 |
ENSRNOT00000078661
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr10_+_110346453 | 0.04 |
ENSRNOT00000054928
|
Tex19.1
|
testis expressed 19.1 |
| chr10_+_95706685 | 0.04 |
ENSRNOT00000004283
|
Psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr2_-_84531192 | 0.04 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr13_-_91872954 | 0.04 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr4_+_163293724 | 0.04 |
ENSRNOT00000077356
|
Gabarapl1
|
GABA type A receptor associated protein like 1 |
| chrX_+_77885586 | 0.04 |
ENSRNOT00000034329
|
RGD1560821
|
similar to small nuclear ribonucleoparticle-associated protein |
| chr3_-_2456041 | 0.04 |
ENSRNOT00000062001
|
Rnf224
|
ring finger protein 224 |
| chrX_-_54303729 | 0.04 |
ENSRNOT00000087919
ENSRNOT00000064340 ENSRNOT00000051249 ENSRNOT00000087547 |
Gk
|
glycerol kinase |
| chr3_-_159802952 | 0.04 |
ENSRNOT00000011610
|
Oser1
|
oxidative stress responsive serine-rich 1 |
| chr4_+_181243338 | 0.04 |
ENSRNOT00000034051
|
Smco2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr4_+_3043231 | 0.04 |
ENSRNOT00000013732
|
Il6
|
interleukin 6 |
| chr3_-_175426395 | 0.04 |
ENSRNOT00000082686
|
Psma7
|
proteasome subunit alpha 7 |
| chr12_+_24761210 | 0.04 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr9_+_38297322 | 0.04 |
ENSRNOT00000078157
ENSRNOT00000088824 |
Bend6
|
BEN domain containing 6 |
| chr2_-_187914905 | 0.04 |
ENSRNOT00000026974
|
Lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr1_-_78997869 | 0.04 |
ENSRNOT00000023490
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr8_-_79715284 | 0.04 |
ENSRNOT00000088562
|
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr10_-_108425206 | 0.04 |
ENSRNOT00000073140
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr16_-_14382641 | 0.04 |
ENSRNOT00000018723
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr5_+_57947716 | 0.03 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chr8_-_132027006 | 0.03 |
ENSRNOT00000050963
|
LOC367195
|
similar to 60S RIBOSOMAL PROTEIN L7 |
| chr4_-_55740627 | 0.03 |
ENSRNOT00000010658
|
Pax4
|
paired box 4 |
| chr2_-_33920837 | 0.03 |
ENSRNOT00000075196
|
Erbin
|
erbb2 interacting protein |
| chr2_-_196207560 | 0.03 |
ENSRNOT00000028589
|
Psmd4
|
proteasome 26S subunit, non-ATPase 4 |
| chr5_+_154868534 | 0.03 |
ENSRNOT00000091442
|
Luzp1
|
leucine zipper protein 1 |
| chr6_+_76188083 | 0.03 |
ENSRNOT00000009666
|
Psma6
|
proteasome subunit alpha 6 |
| chr5_-_159293673 | 0.03 |
ENSRNOT00000009081
|
Padi4
|
peptidyl arginine deiminase 4 |
| chr4_-_9881484 | 0.03 |
ENSRNOT00000016450
|
Psmc2
|
proteasome 26S subunit, ATPase 2 |
| chr7_+_2479311 | 0.03 |
ENSRNOT00000003787
|
Ptges3
|
prostaglandin E synthase 3 |
| chr2_+_60180215 | 0.03 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr11_+_86797557 | 0.03 |
ENSRNOT00000083049
ENSRNOT00000046594 |
Tango2
|
transport and golgi organization 2 homolog |
| chrX_+_28593405 | 0.03 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr16_+_23317953 | 0.03 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr18_-_31109025 | 0.03 |
ENSRNOT00000026381
|
Fchsd1
|
FCH and double SH3 domains 1 |
| chr12_+_39554171 | 0.03 |
ENSRNOT00000024347
|
Atp2a2
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 |
| chr18_-_76984203 | 0.03 |
ENSRNOT00000089122
|
Ctdp1
|
CTD phosphatase subunit 1 |
| chr3_+_112242270 | 0.03 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr10_-_65780349 | 0.03 |
ENSRNOT00000013220
|
Tmem199
|
transmembrane protein 199 |
| chr1_-_225128740 | 0.03 |
ENSRNOT00000026897
|
Rom1
|
retinal outer segment membrane protein 1 |
| chr3_+_151285249 | 0.03 |
ENSRNOT00000055254
|
Procr
|
protein C receptor |
| chr1_+_72635267 | 0.03 |
ENSRNOT00000090636
|
Il11
|
interleukin 11 |
| chr19_-_26053762 | 0.03 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr10_-_29367865 | 0.03 |
ENSRNOT00000005304
|
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr8_+_116324040 | 0.03 |
ENSRNOT00000081353
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr11_-_76888178 | 0.03 |
ENSRNOT00000049841
|
Ostn
|
osteocrin |
| chr3_+_4282236 | 0.02 |
ENSRNOT00000042201
|
LOC100364457
|
ribosomal protein L9-like |
| chr19_-_19376789 | 0.02 |
ENSRNOT00000065103
|
Nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr12_+_16988136 | 0.02 |
ENSRNOT00000078925
ENSRNOT00000036744 |
Micall2
|
MICAL-like 2 |
| chr5_+_144701949 | 0.02 |
ENSRNOT00000015618
|
Psmb2
|
proteasome subunit beta 2 |
| chr7_-_127058056 | 0.02 |
ENSRNOT00000022882
|
Cerk
|
ceramide kinase |
| chr10_+_43224269 | 0.02 |
ENSRNOT00000003473
|
Sap30l
|
SAP30-like |
| chr11_+_82415995 | 0.02 |
ENSRNOT00000051772
|
AABR07034641.1
|
|
| chr11_-_83944763 | 0.02 |
ENSRNOT00000002358
|
Psmd2
|
proteasome 26S subunit, non-ATPase 2 |
| chr17_+_53102534 | 0.02 |
ENSRNOT00000021433
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr13_+_60545635 | 0.02 |
ENSRNOT00000077153
|
Uchl5
|
ubiquitin C-terminal hydrolase L5 |
| chr4_+_176994129 | 0.02 |
ENSRNOT00000018734
|
Cmas
|
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr8_-_13290773 | 0.02 |
ENSRNOT00000012217
|
RGD1561795
|
similar to RIKEN cDNA 1700012B09 |
| chr5_+_59063531 | 0.02 |
ENSRNOT00000085311
ENSRNOT00000065909 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr3_+_79876938 | 0.02 |
ENSRNOT00000015757
|
Psmc3
|
proteasome 26S subunit, ATPase 3 |
| chrX_+_15321713 | 0.02 |
ENSRNOT00000071835
|
LOC108348138
|
GTPase ERas |
| chr6_+_128073344 | 0.02 |
ENSRNOT00000014073
|
LOC500712
|
Ab1-233 |
| chr16_-_755990 | 0.02 |
ENSRNOT00000013501
|
Polr3a
|
RNA polymerase III subunit A |
| chr18_-_60002529 | 0.02 |
ENSRNOT00000081014
ENSRNOT00000024264 ENSRNOT00000059162 |
Nars
|
asparaginyl-tRNA synthetase |
| chr2_+_3662763 | 0.02 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr8_+_5522739 | 0.02 |
ENSRNOT00000011507
|
Mmp13
|
matrix metallopeptidase 13 |
| chr15_-_106844426 | 0.02 |
ENSRNOT00000015890
|
Slc15a1
|
solute carrier family 15 member 1 |
| chr13_-_99287887 | 0.02 |
ENSRNOT00000004780
|
Ephx1
|
epoxide hydrolase 1 |
| chr10_-_56412544 | 0.02 |
ENSRNOT00000020578
|
Tmem102
|
transmembrane protein 102 |
| chr15_+_8730871 | 0.02 |
ENSRNOT00000081845
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chrX_-_107542510 | 0.02 |
ENSRNOT00000074140
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr19_-_39646693 | 0.02 |
ENSRNOT00000019104
|
Psmd7
|
proteasome 26S subunit, non-ATPase 7 |
| chr3_+_175548174 | 0.02 |
ENSRNOT00000091941
|
Adrm1
|
adhesion regulating molecule 1 |
| chr11_+_66713888 | 0.02 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr1_-_175676699 | 0.02 |
ENSRNOT00000030474
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr3_+_138508899 | 0.02 |
ENSRNOT00000009437
|
Kat14
|
lysine acetyltransferase 14 |
| chr18_-_59975192 | 0.02 |
ENSRNOT00000024328
|
Fech
|
ferrochelatase |
| chr19_+_52566607 | 0.02 |
ENSRNOT00000022432
|
Usp10
|
ubiquitin specific peptidase 10 |
| chr9_-_113328428 | 0.02 |
ENSRNOT00000086340
|
Vapa
|
VAMP associated protein A |
| chr9_+_93080615 | 0.02 |
ENSRNOT00000024306
|
Psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr3_+_2490518 | 0.02 |
ENSRNOT00000015099
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr1_+_64740487 | 0.02 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr1_-_183744586 | 0.02 |
ENSRNOT00000015946
|
Psma1
|
proteasome subunit alpha 1 |
| chr13_-_51076852 | 0.02 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr10_-_88019967 | 0.02 |
ENSRNOT00000047580
|
Krt36
|
keratin 36 |
| chr7_+_11152038 | 0.02 |
ENSRNOT00000006168
|
Nfic
|
nuclear factor I/C |
| chr1_+_31531143 | 0.02 |
ENSRNOT00000034704
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr10_+_46906115 | 0.02 |
ENSRNOT00000034006
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr6_+_69950500 | 0.02 |
ENSRNOT00000049500
|
RGD1559629
|
similar to H+ ATP synthase |
| chr1_-_100671074 | 0.02 |
ENSRNOT00000027132
|
Myh14
|
myosin heavy chain 14 |
| chr15_-_61564695 | 0.02 |
ENSRNOT00000068216
|
Rgcc
|
regulator of cell cycle |
| chr6_-_86822094 | 0.01 |
ENSRNOT00000006531
|
Fkbp3
|
FK506 binding protein 3 |
| chr1_-_57489727 | 0.01 |
ENSRNOT00000002037
|
Psmb1
|
proteasome subunit beta 1 |
| chr12_+_40553741 | 0.01 |
ENSRNOT00000057396
|
LOC100360238
|
rCG21419-like |
| chr14_-_33031282 | 0.01 |
ENSRNOT00000043244
|
LOC103693375
|
60S ribosomal protein L39 |
| chr3_+_103747654 | 0.01 |
ENSRNOT00000006887
|
Nop10
|
NOP10 ribonucleoprotein |
| chr5_-_144341586 | 0.01 |
ENSRNOT00000014586
|
Adprhl2
|
ADP-ribosylhydrolase like 2 |
| chr5_-_58445953 | 0.01 |
ENSRNOT00000046102
|
Vcp
|
valosin-containing protein |
| chr7_-_115963046 | 0.01 |
ENSRNOT00000007923
|
Slurp1
|
secreted Ly6/Plaur domain containing 1 |
| chr17_+_6840463 | 0.01 |
ENSRNOT00000061233
|
Ubqln1
|
ubiquilin 1 |
| chr2_+_198040536 | 0.01 |
ENSRNOT00000028744
|
Anp32e
|
acidic nuclear phosphoprotein 32 family member E |
| chr9_+_119353840 | 0.01 |
ENSRNOT00000085362
|
Myom1
|
myomesin 1 |
| chr3_-_79892429 | 0.01 |
ENSRNOT00000016191
|
Slc39a13
|
solute carrier family 39 member 13 |
| chr1_-_242373764 | 0.01 |
ENSRNOT00000076544
|
Fam122a
|
family with sequence similarity 122A |
| chr3_+_57817693 | 0.01 |
ENSRNOT00000067729
ENSRNOT00000047243 ENSRNOT00000013400 |
Dync1i2
|
dynein cytoplasmic 1 intermediate chain 2 |
| chr20_+_5040337 | 0.01 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr13_+_107580629 | 0.01 |
ENSRNOT00000003557
|
LOC100361993
|
transmembrane protein 14C |
| chr1_+_205911552 | 0.01 |
ENSRNOT00000024751
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr10_+_94445877 | 0.01 |
ENSRNOT00000013997
|
Psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr2_+_30347000 | 0.01 |
ENSRNOT00000024291
|
Serf1
|
small EDRK-rich factor 1 |
| chr2_-_187258140 | 0.01 |
ENSRNOT00000017735
|
Prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr15_-_51400606 | 0.01 |
ENSRNOT00000023318
|
Chmp7
|
charged multivesicular body protein 7 |
| chr3_+_146365092 | 0.01 |
ENSRNOT00000008865
|
Cst7
|
cystatin F |
| chr1_+_277068761 | 0.01 |
ENSRNOT00000044183
ENSRNOT00000022382 |
Habp2
|
hyaluronan binding protein 2 |
| chrX_+_159158194 | 0.01 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr1_+_282568287 | 0.01 |
ENSRNOT00000015997
|
Ces2i
|
carboxylesterase 2I |
| chr1_-_252461461 | 0.01 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr12_-_22748860 | 0.01 |
ENSRNOT00000001923
|
Cldn15
|
claudin 15 |
| chrX_-_124464963 | 0.01 |
ENSRNOT00000036472
ENSRNOT00000077697 |
Tmem255a
|
transmembrane protein 255A |
| chr8_-_94368834 | 0.01 |
ENSRNOT00000078977
|
Me1
|
malic enzyme 1 |
| chr8_+_116307141 | 0.01 |
ENSRNOT00000037375
|
Rassf1
|
Ras association domain family member 1 |
| chr8_+_80965255 | 0.01 |
ENSRNOT00000079508
|
Wdr72
|
WD repeat domain 72 |
| chr8_-_130550388 | 0.01 |
ENSRNOT00000026355
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1 |
| chr9_-_85243001 | 0.01 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr2_+_211050360 | 0.01 |
ENSRNOT00000026928
|
Psma5
|
proteasome subunit alpha 5 |
| chr12_-_8759599 | 0.01 |
ENSRNOT00000073155
|
Pomp
|
proteasome maturation protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2_Bach1_Mafk
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 0.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.2 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 0.0 | 0.1 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.1 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.0 | 0.1 | GO:0048619 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.0 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:1990168 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:1904178 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.0 | 0.0 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.3 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.0 | GO:2000660 | negative regulation of chemokine biosynthetic process(GO:0045079) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:1904569 | response to selenite ion(GO:0072714) regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.0 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |