Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Nfatc3
Z-value: 0.53
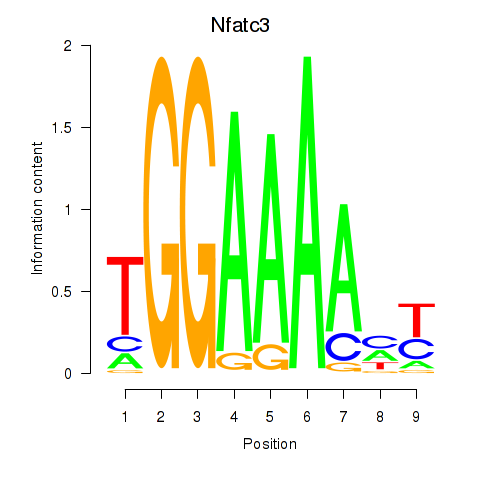
Transcription factors associated with Nfatc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc3
|
ENSRNOG00000054264 | nuclear factor of activated T-cells 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc3 | rn6_v1_chr19_+_38039564_38039564 | 0.28 | 6.4e-01 | Click! |
Activity profile of Nfatc3 motif
Sorted Z-values of Nfatc3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_138148967 | 0.23 |
ENSRNOT00000033968
|
Frmd7
|
FERM domain containing 7 |
| chr3_-_107760550 | 0.19 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr12_-_51965779 | 0.19 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr8_+_127144903 | 0.18 |
ENSRNOT00000013530
|
Eomes
|
eomesodermin |
| chr10_-_74769637 | 0.18 |
ENSRNOT00000008889
|
LOC102555183
|
zinc finger protein OZF-like |
| chr16_+_35573058 | 0.18 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr10_-_32471454 | 0.18 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
| chr17_-_51912496 | 0.18 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr18_+_30890869 | 0.15 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr14_-_112946204 | 0.15 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr18_-_77322690 | 0.15 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr2_-_140618405 | 0.15 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_+_224882439 | 0.15 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr16_-_74020437 | 0.14 |
ENSRNOT00000037389
|
Kat6a
|
lysine acetyltransferase 6A |
| chr10_-_103687425 | 0.13 |
ENSRNOT00000039284
|
Cd300lf
|
Cd300 molecule-like family member F |
| chr1_-_2017488 | 0.13 |
ENSRNOT00000021905
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr14_+_104821215 | 0.12 |
ENSRNOT00000007162
|
Sertad2
|
SERTA domain containing 2 |
| chr1_-_197568165 | 0.12 |
ENSRNOT00000079044
|
Xpo6
|
exportin 6 |
| chr4_-_126071261 | 0.12 |
ENSRNOT00000080234
|
LOC100361920
|
dynein light chain 1-like |
| chr18_+_30562178 | 0.12 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr13_+_70379346 | 0.12 |
ENSRNOT00000038183
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr13_+_105684420 | 0.12 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr4_-_125808281 | 0.12 |
ENSRNOT00000037848
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr4_-_145147397 | 0.11 |
ENSRNOT00000010347
|
Lhfpl4
|
lipoma HMGIC fusion partner-like 4 |
| chr8_-_45137893 | 0.11 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr18_+_30909490 | 0.11 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr6_-_107325345 | 0.11 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr8_-_82533689 | 0.11 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr10_-_16046033 | 0.11 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_+_56627411 | 0.11 |
ENSRNOT00000089108
ENSRNOT00000068493 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr1_-_222177421 | 0.11 |
ENSRNOT00000078393
|
Esrra
|
estrogen related receptor, alpha |
| chr11_-_71135493 | 0.11 |
ENSRNOT00000050535
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr3_+_80556668 | 0.10 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr7_-_83701447 | 0.10 |
ENSRNOT00000088893
|
AABR07057675.1
|
|
| chr4_+_96562725 | 0.10 |
ENSRNOT00000009094
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr5_+_173256637 | 0.10 |
ENSRNOT00000025531
|
Ccnl2
|
cyclin L2 |
| chr3_+_80676820 | 0.10 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr10_-_86554478 | 0.10 |
ENSRNOT00000049621
|
Ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr10_-_38985466 | 0.10 |
ENSRNOT00000010121
|
Il13
|
interleukin 13 |
| chr12_-_51341663 | 0.10 |
ENSRNOT00000066892
ENSRNOT00000052202 |
Pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr10_+_83655460 | 0.10 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr3_-_7795758 | 0.10 |
ENSRNOT00000045919
|
Ntng2
|
netrin G2 |
| chr3_-_110324084 | 0.10 |
ENSRNOT00000010381
|
Bmf
|
Bcl2 modifying factor |
| chr1_+_213583606 | 0.09 |
ENSRNOT00000088899
|
AC109844.1
|
|
| chr3_+_151508361 | 0.09 |
ENSRNOT00000055251
|
Cep250
|
centrosomal protein 250 |
| chr13_+_82231030 | 0.09 |
ENSRNOT00000003551
|
Scyl3
|
SCY1 like pseudokinase 3 |
| chr5_+_22392732 | 0.09 |
ENSRNOT00000087182
|
Clvs1
|
clavesin 1 |
| chr3_-_94418711 | 0.09 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chrX_+_151103576 | 0.09 |
ENSRNOT00000015401
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr9_-_32019205 | 0.09 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr10_-_83655182 | 0.09 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr18_+_29966245 | 0.09 |
ENSRNOT00000074028
|
Pcdha4
|
protocadherin alpha 4 |
| chr11_+_84745904 | 0.08 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr12_-_35979193 | 0.08 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr18_-_14016713 | 0.08 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr2_+_58448917 | 0.08 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chrX_-_157392054 | 0.08 |
ENSRNOT00000085139
ENSRNOT00000082838 |
Zfp92
|
zinc finger protein 92 |
| chr3_+_149741312 | 0.08 |
ENSRNOT00000070854
|
LOC100911637
|
high mobility group protein B1-like |
| chr20_-_3419831 | 0.07 |
ENSRNOT00000046798
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr17_+_36691875 | 0.07 |
ENSRNOT00000074314
|
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr2_+_186872520 | 0.07 |
ENSRNOT00000021577
ENSRNOT00000093312 |
Etv3
|
ets variant 3 |
| chr7_+_15279611 | 0.07 |
ENSRNOT00000043826
|
Zfp952
|
zinc finger protein 952 |
| chr4_-_154044493 | 0.07 |
ENSRNOT00000076318
|
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chr8_-_96266342 | 0.07 |
ENSRNOT00000078891
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr15_+_96821832 | 0.07 |
ENSRNOT00000012785
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr9_+_60039297 | 0.07 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr18_+_30496318 | 0.07 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr9_+_50892605 | 0.07 |
ENSRNOT00000033133
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr2_+_150756185 | 0.07 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chr3_-_12467662 | 0.07 |
ENSRNOT00000051893
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr13_-_83202864 | 0.07 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr17_+_8135251 | 0.06 |
ENSRNOT00000016982
|
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr19_-_1074333 | 0.06 |
ENSRNOT00000017983
ENSRNOT00000086995 |
Cdh5
|
cadherin 5 |
| chr6_+_83421882 | 0.06 |
ENSRNOT00000084258
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr2_+_187988925 | 0.06 |
ENSRNOT00000093033
ENSRNOT00000093016 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr1_-_57815038 | 0.06 |
ENSRNOT00000075401
|
Rgmb
|
repulsive guidance molecule family member B |
| chr9_-_10462491 | 0.06 |
ENSRNOT00000081770
|
Safb
|
scaffold attachment factor B |
| chr17_-_44639919 | 0.06 |
ENSRNOT00000079676
|
Zfp184
|
zinc finger protein 184 |
| chr16_+_59077574 | 0.06 |
ENSRNOT00000090477
|
Dlc1
|
DLC1 Rho GTPase activating protein |
| chr1_+_66898946 | 0.06 |
ENSRNOT00000074063
|
Zfp551
|
zinc finger protein 551 |
| chr2_-_89498395 | 0.06 |
ENSRNOT00000068507
|
AABR07009224.1
|
|
| chr10_+_49259194 | 0.06 |
ENSRNOT00000091100
ENSRNOT00000004390 |
Fbxw10
|
F-box and WD repeat domain containing 10 |
| chr18_+_29951094 | 0.06 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr10_+_46511271 | 0.06 |
ENSRNOT00000080828
|
Rai1
|
retinoic acid induced 1 |
| chr6_-_3355339 | 0.06 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr1_+_171797516 | 0.06 |
ENSRNOT00000088110
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr6_-_125060482 | 0.06 |
ENSRNOT00000034961
|
Ppp4r3a
|
protein phosphatase 4, regulatory subunit 3A |
| chr10_+_70136855 | 0.06 |
ENSRNOT00000084076
|
Lig3
|
DNA ligase 3 |
| chr16_+_11032531 | 0.05 |
ENSRNOT00000078390
|
Wapl
|
WAPL cohesin release factor |
| chr7_+_25808419 | 0.05 |
ENSRNOT00000091571
|
Rfx4
|
regulatory factor X4 |
| chr7_-_36499784 | 0.05 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr13_-_79744939 | 0.05 |
ENSRNOT00000076706
|
Suco
|
SUN domain containing ossification factor |
| chr7_-_138483612 | 0.05 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr14_+_7618022 | 0.05 |
ENSRNOT00000088508
ENSRNOT00000002819 |
Slc10a6
|
solute carrier family 10 member 6 |
| chr18_+_53915807 | 0.05 |
ENSRNOT00000026543
|
Adamts19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19 |
| chr1_+_37507276 | 0.05 |
ENSRNOT00000047627
|
Adcy2
|
adenylate cyclase 2 |
| chr15_+_106434177 | 0.05 |
ENSRNOT00000015271
|
Farp1
|
FERM, ARH/RhoGEF and pleckstrin domain protein 1 |
| chr9_+_71230108 | 0.05 |
ENSRNOT00000018326
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr3_+_8547349 | 0.05 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr6_+_115170865 | 0.05 |
ENSRNOT00000005671
|
Tshr
|
thyroid stimulating hormone receptor |
| chr4_+_27473477 | 0.05 |
ENSRNOT00000007940
ENSRNOT00000079571 |
Ankib1
|
ankyrin repeat and IBR domain containing 1 |
| chr8_-_107656861 | 0.05 |
ENSRNOT00000050374
|
Mras
|
muscle RAS oncogene homolog |
| chr15_-_34469350 | 0.05 |
ENSRNOT00000067536
|
Adcy4
|
adenylate cyclase 4 |
| chr18_-_76653056 | 0.05 |
ENSRNOT00000078496
|
Adnp2
|
ADNP homeobox 2 |
| chr1_+_164706276 | 0.05 |
ENSRNOT00000024311
|
Olr36
|
olfactory receptor 36 |
| chr1_-_6970040 | 0.05 |
ENSRNOT00000016273
|
Utrn
|
utrophin |
| chrX_-_82743753 | 0.05 |
ENSRNOT00000003512
|
Rps6ka6
|
ribosomal protein S6 kinase A6 |
| chr9_-_44237117 | 0.05 |
ENSRNOT00000068496
|
RGD1310819
|
similar to putative protein (5S487) |
| chr20_+_27129291 | 0.05 |
ENSRNOT00000031053
|
AABR07044933.1
|
|
| chr6_+_69971227 | 0.04 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr4_+_22084954 | 0.04 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr1_+_198682230 | 0.04 |
ENSRNOT00000023995
|
Znf48
|
zinc finger protein 48 |
| chr1_-_131454689 | 0.04 |
ENSRNOT00000014152
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_-_131460473 | 0.04 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr13_+_90116843 | 0.04 |
ENSRNOT00000006306
|
Cd48
|
Cd48 molecule |
| chr2_+_57206613 | 0.04 |
ENSRNOT00000082694
ENSRNOT00000046069 |
Nup155
|
nucleoporin 155 |
| chr20_+_27832932 | 0.04 |
ENSRNOT00000001092
|
Gcc2
|
GRIP and coiled-coil domain containing 2 |
| chr13_-_103080920 | 0.04 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chr10_+_90930010 | 0.04 |
ENSRNOT00000003765
|
Higd1b
|
HIG1 hypoxia inducible domain family, member 1B |
| chr1_-_215310013 | 0.04 |
ENSRNOT00000054858
|
AABR07006049.2
|
|
| chr5_+_173256834 | 0.04 |
ENSRNOT00000089936
|
Ccnl2
|
cyclin L2 |
| chr16_-_10603850 | 0.04 |
ENSRNOT00000087737
|
Fam35a
|
family with sequence similarity 35, member A |
| chr12_-_29743705 | 0.04 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr4_-_123713319 | 0.04 |
ENSRNOT00000012875
ENSRNOT00000086982 |
Slc6a6
|
solute carrier family 6 member 6 |
| chr3_-_71845232 | 0.04 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr2_-_174413038 | 0.04 |
ENSRNOT00000089229
ENSRNOT00000037115 |
Golim4
|
golgi integral membrane protein 4 |
| chr1_+_267359830 | 0.04 |
ENSRNOT00000015495
|
Slk
|
STE20-like kinase |
| chr11_+_82862695 | 0.04 |
ENSRNOT00000071707
ENSRNOT00000073435 |
Clcn2
|
chloride voltage-gated channel 2 |
| chr16_-_71203609 | 0.04 |
ENSRNOT00000088458
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr3_-_162581610 | 0.04 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr2_-_123851854 | 0.04 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr3_+_67849966 | 0.04 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr5_+_152333043 | 0.04 |
ENSRNOT00000021342
|
Ubxn11
|
UBX domain protein 11 |
| chr17_+_55008265 | 0.04 |
ENSRNOT00000033827
|
Zfp438
|
zinc finger protein 438 |
| chr17_-_27665266 | 0.03 |
ENSRNOT00000060218
|
Rreb1
|
ras responsive element binding protein 1 |
| chr1_-_165123455 | 0.03 |
ENSRNOT00000024875
|
Pold3
|
DNA polymerase delta 3, accessory subunit |
| chr1_+_167539036 | 0.03 |
ENSRNOT00000093112
|
Rrm1
|
ribonucleotide reductase catalytic subunit M1 |
| chr2_-_243224883 | 0.03 |
ENSRNOT00000014139
|
Dapp1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides 1 |
| chr18_-_28017925 | 0.03 |
ENSRNOT00000075420
|
Lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr20_-_49486550 | 0.03 |
ENSRNOT00000048270
ENSRNOT00000076541 |
Prdm1
|
PR/SET domain 1 |
| chr14_-_44225713 | 0.03 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr2_-_210088949 | 0.03 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr14_+_79205466 | 0.03 |
ENSRNOT00000085534
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chr3_+_113251778 | 0.03 |
ENSRNOT00000083005
|
Map1a
|
microtubule-associated protein 1A |
| chr4_-_42188289 | 0.03 |
ENSRNOT00000081688
|
Rbmxl1b
|
RNA binding motif protein, X-linked-like 1B |
| chr7_-_132984110 | 0.03 |
ENSRNOT00000046626
|
Hmgb1-ps3
|
high mobility group box 1, pseudogene 3 |
| chr11_+_7265828 | 0.03 |
ENSRNOT00000084765
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr8_+_73682887 | 0.03 |
ENSRNOT00000057522
|
Vps13c
|
vacuolar protein sorting 13C |
| chr14_+_5928737 | 0.03 |
ENSRNOT00000071877
ENSRNOT00000040985 ENSRNOT00000074889 |
Mpa2l
|
macrophage activation 2 like |
| chr6_-_129509720 | 0.03 |
ENSRNOT00000006131
|
Atg2b
|
autophagy related 2B |
| chr15_-_56970365 | 0.03 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr4_+_180291389 | 0.03 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr17_-_44640092 | 0.02 |
ENSRNOT00000077628
|
Zfp184
|
zinc finger protein 184 |
| chr2_-_18531210 | 0.02 |
ENSRNOT00000088313
|
Vcan
|
versican |
| chr10_+_63803309 | 0.02 |
ENSRNOT00000036666
|
Myo1c
|
myosin 1C |
| chr13_-_73819896 | 0.02 |
ENSRNOT00000036392
|
Fam163a
|
family with sequence similarity 163, member A |
| chrX_+_115351114 | 0.02 |
ENSRNOT00000047553
|
Hmg1l1
|
high-mobility group (nonhistone chromosomal) protein 1-like 1 |
| chr13_+_89586283 | 0.02 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_-_117267803 | 0.02 |
ENSRNOT00000082271
|
Plec
|
plectin |
| chr4_-_170932618 | 0.02 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr4_-_56114254 | 0.02 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr1_-_266914093 | 0.02 |
ENSRNOT00000027526
|
Calhm2
|
calcium homeostasis modulator 2 |
| chr14_+_34455934 | 0.02 |
ENSRNOT00000085991
ENSRNOT00000002981 |
Clock
|
clock circadian regulator |
| chr5_+_64294321 | 0.02 |
ENSRNOT00000083796
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr3_-_48535909 | 0.02 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr10_+_52334240 | 0.02 |
ENSRNOT00000005518
|
Zfp18
|
zinc finger protein 18 |
| chrX_+_17540458 | 0.02 |
ENSRNOT00000045710
|
Nudt11
|
nudix hydrolase 11 |
| chr5_+_61657507 | 0.02 |
ENSRNOT00000013228
|
Tmod1
|
tropomodulin 1 |
| chr11_+_61609370 | 0.02 |
ENSRNOT00000088880
ENSRNOT00000082533 |
Gramd1c
|
GRAM domain containing 1C |
| chr16_-_6404578 | 0.02 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr2_+_243502073 | 0.02 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_-_62128044 | 0.02 |
ENSRNOT00000093141
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr1_-_211196868 | 0.02 |
ENSRNOT00000022714
|
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr8_+_64610952 | 0.02 |
ENSRNOT00000015963
|
Myo9a
|
myosin IXA |
| chr10_-_78690857 | 0.02 |
ENSRNOT00000089255
|
LOC303448
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr13_-_89606326 | 0.02 |
ENSRNOT00000029179
|
Fcer1g
|
Fc fragment of IgE receptor Ig |
| chr15_+_4285052 | 0.02 |
ENSRNOT00000035345
|
Cfap70
|
cilia and flagella associated protein 70 |
| chr20_+_27366213 | 0.02 |
ENSRNOT00000000302
ENSRNOT00000057939 |
Tet1
|
tet methylcytosine dioxygenase 1 |
| chr16_-_73827488 | 0.02 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr7_-_29701586 | 0.02 |
ENSRNOT00000009084
ENSRNOT00000089269 |
Ano4
|
anoctamin 4 |
| chr15_+_39779648 | 0.01 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr3_-_105298108 | 0.01 |
ENSRNOT00000010994
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr8_-_114449956 | 0.01 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr11_+_61761613 | 0.01 |
ENSRNOT00000089340
ENSRNOT00000091598 |
Qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr1_+_174702373 | 0.01 |
ENSRNOT00000013733
|
Zfp143
|
zinc finger protein 143 |
| chr11_-_64421248 | 0.01 |
ENSRNOT00000066997
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr16_-_3912148 | 0.01 |
ENSRNOT00000014941
|
Anxa11
|
annexin A11 |
| chr6_+_99433550 | 0.01 |
ENSRNOT00000079359
ENSRNOT00000008504 |
Hspa2
|
heat shock protein family A member 2 |
| chr10_-_55924158 | 0.01 |
ENSRNOT00000011185
|
Cntrob
|
centrobin, centriole duplication and spindle assembly protein |
| chr10_+_71322145 | 0.01 |
ENSRNOT00000079418
|
Synrg
|
synergin, gamma |
| chr4_-_176922424 | 0.01 |
ENSRNOT00000055540
|
Abcc9
|
ATP binding cassette subfamily C member 9 |
| chr2_-_198120041 | 0.01 |
ENSRNOT00000079621
ENSRNOT00000028747 |
Plekho1
|
pleckstrin homology domain containing O1 |
| chr1_+_1771710 | 0.01 |
ENSRNOT00000080138
ENSRNOT00000073528 |
Nup43
|
nucleoporin 43 |
| chr8_+_59507990 | 0.01 |
ENSRNOT00000018003
|
Hykk
|
hydroxylysine kinase |
| chr20_+_4959294 | 0.01 |
ENSRNOT00000074223
|
Hspa1l
|
heat shock protein family A (Hsp70) member 1 like |
| chr7_-_117267402 | 0.01 |
ENSRNOT00000088945
|
Plec
|
plectin |
| chr2_-_29121104 | 0.01 |
ENSRNOT00000020543
|
Tnpo1
|
transportin 1 |
| chr18_+_29987206 | 0.01 |
ENSRNOT00000027383
|
Pcdha4
|
protocadherin alpha 4 |
| chr15_-_70349983 | 0.01 |
ENSRNOT00000012167
|
Diaph3
|
diaphanous-related formin 3 |
| chr1_-_6903486 | 0.01 |
ENSRNOT00000086574
|
Utrn
|
utrophin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.0 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
| 0.0 | 0.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0008458 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.0 | 0.0 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.0 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |