Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Nfatc1
Z-value: 1.07
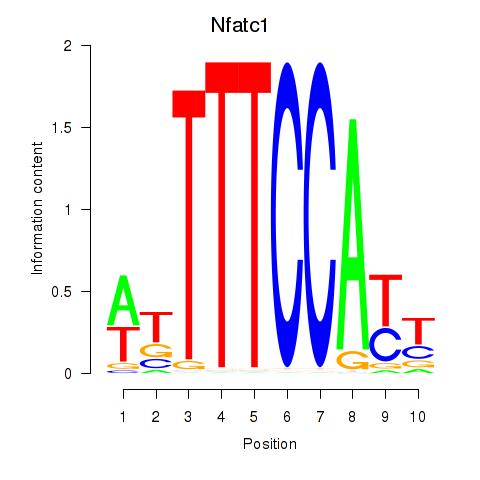
Transcription factors associated with Nfatc1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc1
|
ENSRNOG00000017146 | nuclear factor of activated T-cells 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | rn6_v1_chr18_-_77322690_77322690 | 0.59 | 2.9e-01 | Click! |
Activity profile of Nfatc1 motif
Sorted Z-values of Nfatc1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_137934484 | 0.75 |
ENSRNOT00000049046
|
LOC317588
|
hypothetical protein LOC317588 |
| chr20_-_22459025 | 0.66 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr13_+_29839867 | 0.60 |
ENSRNOT00000090623
|
AABR07020537.1
|
|
| chr2_+_149899836 | 0.50 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr13_+_41802697 | 0.47 |
ENSRNOT00000046719
|
LOC100362110
|
LRRGT00155-like |
| chr1_-_4560789 | 0.46 |
ENSRNOT00000082415
|
Adgb
|
androglobin |
| chr1_+_168668707 | 0.46 |
ENSRNOT00000015108
|
AC107531.3
|
|
| chr17_+_48436720 | 0.44 |
ENSRNOT00000074309
|
AABR07027902.1
|
|
| chr1_+_1545134 | 0.44 |
ENSRNOT00000044523
|
AABR07000156.1
|
|
| chr2_-_112368557 | 0.44 |
ENSRNOT00000075017
|
AABR07009787.1
|
|
| chr7_-_99808612 | 0.41 |
ENSRNOT00000074953
|
AABR07058091.2
|
|
| chr10_-_36419926 | 0.41 |
ENSRNOT00000004902
|
Znf354b
|
zinc finger protein 354B |
| chr1_+_254531659 | 0.40 |
ENSRNOT00000091377
|
AC080157.1
|
|
| chr2_+_118746333 | 0.37 |
ENSRNOT00000079431
|
AABR07009965.1
|
|
| chr3_+_95614562 | 0.36 |
ENSRNOT00000079990
|
AABR07053179.1
|
|
| chr2_+_70490155 | 0.34 |
ENSRNOT00000078674
|
AABR07008681.1
|
|
| chr2_+_11658568 | 0.33 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr18_-_77322690 | 0.33 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr12_-_13374500 | 0.33 |
ENSRNOT00000038577
|
Zfp12
|
zinc finger protein 12 |
| chr10_+_83655460 | 0.30 |
ENSRNOT00000008011
|
Gngt2
|
G protein subunit gamma transducin 2 |
| chr3_-_123849788 | 0.29 |
ENSRNOT00000028869
|
Rnf24
|
ring finger protein 24 |
| chr2_+_216863428 | 0.28 |
ENSRNOT00000068413
|
Col11a1
|
collagen type XI alpha 1 chain |
| chr14_+_39663421 | 0.28 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr8_-_84632817 | 0.28 |
ENSRNOT00000076942
|
Mlip
|
muscular LMNA-interacting protein |
| chr4_-_157433467 | 0.27 |
ENSRNOT00000028965
|
Lag3
|
lymphocyte activating 3 |
| chr5_-_8864578 | 0.27 |
ENSRNOT00000008480
|
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr5_+_27326762 | 0.27 |
ENSRNOT00000066191
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr13_+_67351087 | 0.26 |
ENSRNOT00000003567
|
Ptgs2
|
prostaglandin-endoperoxide synthase 2 |
| chr7_-_98098268 | 0.26 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chrX_-_31851715 | 0.25 |
ENSRNOT00000068601
|
Vegfd
|
vascular endothelial growth factor D |
| chr18_-_1389929 | 0.25 |
ENSRNOT00000051362
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr18_-_1390190 | 0.24 |
ENSRNOT00000061777
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1 |
| chr7_+_48867664 | 0.23 |
ENSRNOT00000005862
|
Ppfia2
|
PTPRF interacting protein alpha 2 |
| chr18_+_30864216 | 0.23 |
ENSRNOT00000027015
|
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chrX_+_114929029 | 0.23 |
ENSRNOT00000006459
|
Pak3
|
p21 (RAC1) activated kinase 3 |
| chr1_-_62316450 | 0.22 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chr1_-_266914093 | 0.22 |
ENSRNOT00000027526
|
Calhm2
|
calcium homeostasis modulator 2 |
| chr2_+_54191538 | 0.22 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr14_+_19866408 | 0.21 |
ENSRNOT00000060465
|
Adamts3
|
ADAM metallopeptidase with thrombospondin type 1, motif 3 |
| chr11_+_61531416 | 0.21 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr1_-_128341240 | 0.21 |
ENSRNOT00000070864
ENSRNOT00000072915 |
Mef2a
|
myocyte enhancer factor 2a |
| chr1_+_88885937 | 0.21 |
ENSRNOT00000029719
|
Nfkbid
|
NFKB inhibitor delta |
| chr1_+_44311513 | 0.20 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr9_+_43889473 | 0.20 |
ENSRNOT00000024330
|
Inpp4a
|
inositol polyphosphate-4-phosphatase type I A |
| chr4_+_120672152 | 0.20 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr7_-_134495835 | 0.20 |
ENSRNOT00000058349
|
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr5_-_160352927 | 0.20 |
ENSRNOT00000017247
|
Dnajc16
|
DnaJ heat shock protein family (Hsp40) member C16 |
| chr9_-_81772851 | 0.19 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr14_+_109533792 | 0.19 |
ENSRNOT00000067690
|
LOC108348154
|
ankyrin repeat domain-containing protein 29-like |
| chr18_+_65155685 | 0.19 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr13_-_113817995 | 0.19 |
ENSRNOT00000057151
|
Cd46
|
CD46 molecule |
| chr3_+_58164931 | 0.19 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chrY_+_327166 | 0.18 |
ENSRNOT00000092902
|
Sry
|
sex determining region Y |
| chr9_+_67699379 | 0.18 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr16_-_70705128 | 0.18 |
ENSRNOT00000071258
|
Rnf170
|
ring finger protein 170 |
| chr13_-_82005741 | 0.18 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr3_-_64024205 | 0.17 |
ENSRNOT00000037015
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr17_-_27665266 | 0.17 |
ENSRNOT00000060218
|
Rreb1
|
ras responsive element binding protein 1 |
| chr13_-_67688477 | 0.17 |
ENSRNOT00000068148
|
Prg4
|
proteoglycan 4 |
| chr15_-_33250546 | 0.17 |
ENSRNOT00000017857
|
RGD1565222
|
similar to RIKEN cDNA 4931414P19 |
| chr2_+_22910236 | 0.17 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr10_+_34489940 | 0.17 |
ENSRNOT00000085975
|
Zfp62
|
zinc finger protein 62 |
| chr12_-_19294888 | 0.17 |
ENSRNOT00000001815
|
Zfp113
|
zinc finger protein 3 |
| chr16_+_70859280 | 0.17 |
ENSRNOT00000019630
|
Pomk
|
protein-O-mannose kinase |
| chr1_+_224998172 | 0.17 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr5_-_59647633 | 0.17 |
ENSRNOT00000091944
|
Rnf38
|
ring finger protein 38 |
| chr1_+_255185629 | 0.17 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chr4_-_42188289 | 0.16 |
ENSRNOT00000081688
|
Rbmxl1b
|
RNA binding motif protein, X-linked-like 1B |
| chr8_+_2604962 | 0.16 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chrX_+_18163358 | 0.16 |
ENSRNOT00000045517
|
LOC367746
|
similar to Spindlin-like protein 2 (SPIN-2) |
| chr18_+_29386809 | 0.16 |
ENSRNOT00000082079
ENSRNOT00000024825 |
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr2_-_183031214 | 0.16 |
ENSRNOT00000013260
|
LOC679811
|
similar to RIKEN cDNA D930015E06 |
| chr16_+_35116440 | 0.16 |
ENSRNOT00000074353
|
AABR07025358.1
|
|
| chr9_+_73433252 | 0.16 |
ENSRNOT00000092540
|
Map2
|
microtubule-associated protein 2 |
| chr12_-_29743705 | 0.16 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr17_-_51912496 | 0.16 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr2_+_150756185 | 0.16 |
ENSRNOT00000088461
ENSRNOT00000036808 |
Mbnl1
|
muscleblind-like splicing regulator 1 |
| chr8_+_99880073 | 0.16 |
ENSRNOT00000010765
|
Plscr4
|
phospholipid scramblase 4 |
| chr5_-_59553416 | 0.16 |
ENSRNOT00000090490
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr5_-_160383782 | 0.16 |
ENSRNOT00000018349
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr5_-_160070916 | 0.16 |
ENSRNOT00000067517
ENSRNOT00000055791 |
Spen
|
spen family transcriptional repressor |
| chr1_+_190681094 | 0.15 |
ENSRNOT00000036175
|
Vwa3a
|
von Willebrand factor A domain containing 3A |
| chr7_-_36408588 | 0.15 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr5_-_60191941 | 0.15 |
ENSRNOT00000033373
|
Pax5
|
paired box 5 |
| chr3_+_8451292 | 0.15 |
ENSRNOT00000020407
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr13_-_89545182 | 0.15 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr13_-_73819896 | 0.15 |
ENSRNOT00000036392
|
Fam163a
|
family with sequence similarity 163, member A |
| chr9_-_32019205 | 0.15 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr1_+_171797516 | 0.14 |
ENSRNOT00000088110
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr2_-_89498395 | 0.14 |
ENSRNOT00000068507
|
AABR07009224.1
|
|
| chr16_+_70778913 | 0.14 |
ENSRNOT00000019281
|
Hook3
|
hook microtubule-tethering protein 3 |
| chr3_-_71845232 | 0.14 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr18_+_70427007 | 0.14 |
ENSRNOT00000087959
ENSRNOT00000019512 |
Myo5b
|
myosin Vb |
| chr1_+_213583606 | 0.14 |
ENSRNOT00000088899
|
AC109844.1
|
|
| chrX_-_38196060 | 0.14 |
ENSRNOT00000006741
ENSRNOT00000006438 |
Sh3kbp1
|
SH3 domain-containing kinase-binding protein 1 |
| chr6_-_107325345 | 0.14 |
ENSRNOT00000049481
ENSRNOT00000042594 ENSRNOT00000013026 |
Numb
|
NUMB, endocytic adaptor protein |
| chr7_+_40217991 | 0.13 |
ENSRNOT00000085684
|
Cep290
|
centrosomal protein 290 |
| chr18_+_30036887 | 0.13 |
ENSRNOT00000077824
|
Pcdha4
|
protocadherin alpha 4 |
| chr8_-_39551700 | 0.13 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr11_-_71128299 | 0.13 |
ENSRNOT00000080568
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr9_-_98597359 | 0.13 |
ENSRNOT00000027506
|
Per2
|
period circadian clock 2 |
| chr8_+_76400341 | 0.13 |
ENSRNOT00000087382
ENSRNOT00000081072 |
Bnip2
|
BCL2/adenovirus E1B interacting protein 2 |
| chr1_+_228684136 | 0.13 |
ENSRNOT00000028608
|
Olr337
|
olfactory receptor 337 |
| chr5_-_58484900 | 0.13 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr15_+_39779648 | 0.13 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr2_-_39007976 | 0.12 |
ENSRNOT00000064292
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr6_+_83421882 | 0.12 |
ENSRNOT00000084258
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr17_-_44640092 | 0.12 |
ENSRNOT00000077628
|
Zfp184
|
zinc finger protein 184 |
| chr6_-_42616548 | 0.12 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr5_-_173653905 | 0.12 |
ENSRNOT00000038556
|
Plekhn1
|
pleckstrin homology domain containing N1 |
| chr5_+_150833819 | 0.12 |
ENSRNOT00000056170
|
Eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr5_-_147761983 | 0.12 |
ENSRNOT00000012936
|
Lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr17_+_53231343 | 0.12 |
ENSRNOT00000021703
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr12_-_41266430 | 0.12 |
ENSRNOT00000001853
ENSRNOT00000081108 |
Oas1b
|
2-5 oligoadenylate synthetase 1B |
| chr7_+_130532435 | 0.11 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr5_+_120498883 | 0.11 |
ENSRNOT00000007859
|
Leprot
|
leptin receptor overlapping transcript |
| chr5_+_74766636 | 0.11 |
ENSRNOT00000030913
|
Rn50_5_0791.1
|
|
| chr4_-_124338176 | 0.11 |
ENSRNOT00000016628
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr3_+_170550314 | 0.11 |
ENSRNOT00000006991
|
Tfap2c
|
transcription factor AP-2 gamma |
| chr10_+_95770154 | 0.11 |
ENSRNOT00000030300
|
Helz
|
helicase with zinc finger |
| chr11_-_65208837 | 0.11 |
ENSRNOT00000003867
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr17_+_36334147 | 0.11 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr10_+_49259194 | 0.11 |
ENSRNOT00000091100
ENSRNOT00000004390 |
Fbxw10
|
F-box and WD repeat domain containing 10 |
| chr18_+_30890869 | 0.11 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr1_+_190462327 | 0.11 |
ENSRNOT00000030732
|
LOC691519
|
similar to ankyrin repeat domain 26 |
| chr15_-_57805184 | 0.11 |
ENSRNOT00000000168
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr10_-_84847857 | 0.11 |
ENSRNOT00000071495
|
LOC102552549
|
migration and invasion enhancer 1-like |
| chr3_-_67787990 | 0.10 |
ENSRNOT00000064851
|
Nckap1
|
NCK-associated protein 1 |
| chr15_-_33428942 | 0.10 |
ENSRNOT00000019618
ENSRNOT00000092160 |
Slc7a8
|
solute carrier family 7 member 8 |
| chr6_+_137063611 | 0.10 |
ENSRNOT00000017504
|
LOC691485
|
hypothetical protein LOC691485 |
| chr10_+_84669914 | 0.10 |
ENSRNOT00000011665
|
Cbx1
|
chromobox 1 |
| chr2_-_112802073 | 0.10 |
ENSRNOT00000093081
|
Ect2
|
epithelial cell transforming 2 |
| chr1_+_198655742 | 0.10 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr6_+_143901280 | 0.10 |
ENSRNOT00000091884
ENSRNOT00000005648 |
Zfp386
|
zinc finger protein 386 (Kruppel-like) |
| chr18_+_30562178 | 0.10 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr2_+_187893875 | 0.10 |
ENSRNOT00000093014
|
Mex3a
|
mex-3 RNA binding family member A |
| chr5_-_88612626 | 0.10 |
ENSRNOT00000089560
|
Tle1
|
transducin like enhancer of split 1 |
| chr11_+_61661724 | 0.10 |
ENSRNOT00000079423
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr10_+_68588789 | 0.10 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr3_-_150960030 | 0.10 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chr1_+_151439409 | 0.10 |
ENSRNOT00000022060
ENSRNOT00000050639 |
Grm5
|
glutamate metabotropic receptor 5 |
| chr15_+_32811135 | 0.10 |
ENSRNOT00000067689
|
AABR07017902.1
|
|
| chr20_-_27657983 | 0.10 |
ENSRNOT00000057313
|
Fam26e
|
family with sequence similarity 26, member E |
| chr3_-_153246433 | 0.10 |
ENSRNOT00000067748
|
Samhd1
|
SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase 1 |
| chr4_+_22084954 | 0.09 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr18_+_30023828 | 0.09 |
ENSRNOT00000079008
|
Pcdha4
|
protocadherin alpha 4 |
| chr6_+_10348308 | 0.09 |
ENSRNOT00000034991
|
Epas1
|
endothelial PAS domain protein 1 |
| chr10_-_38969501 | 0.09 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr13_+_50164563 | 0.09 |
ENSRNOT00000029533
|
Lax1
|
lymphocyte transmembrane adaptor 1 |
| chr3_+_100786862 | 0.09 |
ENSRNOT00000080190
|
Bdnf
|
brain-derived neurotrophic factor |
| chr7_+_41114697 | 0.09 |
ENSRNOT00000041354
|
Atp2b1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr10_-_83655182 | 0.09 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr1_-_6903486 | 0.09 |
ENSRNOT00000086574
|
Utrn
|
utrophin |
| chr3_+_100787449 | 0.09 |
ENSRNOT00000078543
|
Bdnf
|
brain-derived neurotrophic factor |
| chr6_-_50943488 | 0.09 |
ENSRNOT00000068419
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr13_+_50434886 | 0.09 |
ENSRNOT00000076857
|
Sox13
|
SRY box 13 |
| chr11_+_38727048 | 0.09 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr15_+_41448064 | 0.09 |
ENSRNOT00000019551
|
Sacs
|
sacsin molecular chaperone |
| chr16_+_6962722 | 0.09 |
ENSRNOT00000023330
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr6_+_64789940 | 0.09 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr13_+_50434702 | 0.08 |
ENSRNOT00000032244
|
Sox13
|
SRY box 13 |
| chr11_+_88699222 | 0.08 |
ENSRNOT00000084177
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_-_276308095 | 0.08 |
ENSRNOT00000054681
|
Zdhhc6
|
zinc finger, DHHC-type containing 6 |
| chr10_+_56193856 | 0.08 |
ENSRNOT00000080584
ENSRNOT00000091216 ENSRNOT00000083458 |
Tp53
|
tumor protein p53 |
| chr17_+_89951752 | 0.08 |
ENSRNOT00000057572
ENSRNOT00000091023 |
Abi1
|
abl-interactor 1 |
| chr9_-_105655471 | 0.08 |
ENSRNOT00000014921
|
RGD1560925
|
similar to 2610034M16Rik protein |
| chr7_-_36499784 | 0.08 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr14_-_44225713 | 0.08 |
ENSRNOT00000049161
|
LOC680579
|
similar to ribosomal protein L14 |
| chr1_-_25839198 | 0.08 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr18_+_750776 | 0.08 |
ENSRNOT00000021981
|
Zfp136
|
zinc finger protein 136 |
| chr5_+_142797366 | 0.08 |
ENSRNOT00000031928
|
Mtf1
|
metal-regulatory transcription factor 1 |
| chr8_-_114449956 | 0.08 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr15_+_23777856 | 0.08 |
ENSRNOT00000060847
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr16_+_75145401 | 0.08 |
ENSRNOT00000046821
ENSRNOT00000047761 |
Spag11bl
|
sperm associated antigen 11b-like |
| chr12_+_41200718 | 0.08 |
ENSRNOT00000038426
ENSRNOT00000048450 ENSRNOT00000067176 |
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr4_+_181055662 | 0.08 |
ENSRNOT00000086813
|
Stk38l
|
serine/threonine kinase 38 like |
| chr3_+_129462738 | 0.08 |
ENSRNOT00000077755
|
AABR07053830.1
|
|
| chr1_+_98627372 | 0.08 |
ENSRNOT00000030370
|
Dzf17
|
zinc finger protein 17 |
| chr9_-_44237117 | 0.08 |
ENSRNOT00000068496
|
RGD1310819
|
similar to putative protein (5S487) |
| chr3_+_172692452 | 0.08 |
ENSRNOT00000077193
|
Zfp831
|
zinc finger protein 831 |
| chr5_+_135351816 | 0.08 |
ENSRNOT00000082602
|
Ipp
|
intracisternal A particle-promoted polypeptide |
| chr4_-_125929002 | 0.08 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr3_-_105214989 | 0.08 |
ENSRNOT00000037895
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr2_+_243502073 | 0.08 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr7_+_27620458 | 0.08 |
ENSRNOT00000059532
|
RGD1560034
|
similar to FLJ25323 protein |
| chr1_+_63842277 | 0.07 |
ENSRNOT00000087957
ENSRNOT00000080820 |
Lilrb3a
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3A |
| chr20_+_48212378 | 0.07 |
ENSRNOT00000040519
|
AABR07045454.1
|
|
| chr15_+_104026601 | 0.07 |
ENSRNOT00000013520
ENSRNOT00000083269 ENSRNOT00000093403 |
Cldn10
|
claudin 10 |
| chr10_+_1920529 | 0.07 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr6_-_39363367 | 0.07 |
ENSRNOT00000088687
ENSRNOT00000065531 |
Fam84a
|
family with sequence similarity 84, member A |
| chr4_+_9981958 | 0.07 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr6_+_18880737 | 0.07 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr16_-_81028656 | 0.07 |
ENSRNOT00000091296
|
Dcun1d2
|
defective in cullin neddylation 1 domain containing 2 |
| chr11_+_42945084 | 0.07 |
ENSRNOT00000002292
|
Crybg3
|
crystallin beta-gamma domain containing 3 |
| chr10_-_15928169 | 0.07 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr7_+_11356118 | 0.07 |
ENSRNOT00000041325
|
Atcay
|
ATCAY, caytaxin |
| chr8_-_36374673 | 0.07 |
ENSRNOT00000013763
|
Dcps
|
decapping enzyme, scavenger |
| chr20_-_5153399 | 0.07 |
ENSRNOT00000001133
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr14_-_90864917 | 0.07 |
ENSRNOT00000044737
|
LOC498414
|
LRRGT00197 |
| chr10_+_36589181 | 0.07 |
ENSRNOT00000045596
|
Zfp354a
|
zinc finger protein 354A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.1 | 0.6 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 0.3 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.3 | GO:2001013 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 | 0.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.2 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 0.3 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.2 | GO:0046380 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.4 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.2 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:1904339 | cellular response to iron(II) ion(GO:0071282) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:1903660 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.1 | GO:1900158 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.0 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0033552 | response to vitamin B3(GO:0033552) |
| 0.0 | 0.1 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.2 | GO:2000020 | positive regulation of male gonad development(GO:2000020) |
| 0.0 | 0.2 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:1905075 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.1 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.0 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.0 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.3 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.0 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.1 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 0.0 | 0.0 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.0 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0016414 | carnitine O-octanoyltransferase activity(GO:0008458) O-octanoyltransferase activity(GO:0016414) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.1 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.0 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.7 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |