Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
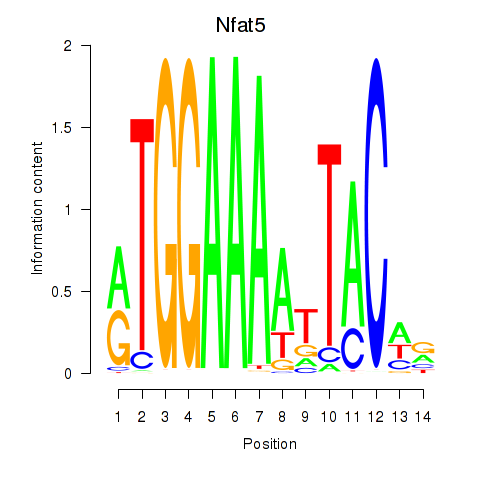
Results for Nfat5
Z-value: 0.31
Transcription factors associated with Nfat5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfat5
|
ENSRNOG00000011879 | nuclear factor of activated T-cells 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfat5 | rn6_v1_chr19_-_38533735_38533735 | 0.51 | 3.8e-01 | Click! |
Activity profile of Nfat5 motif
Sorted Z-values of Nfat5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_218466289 | 0.17 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr17_+_24242642 | 0.07 |
ENSRNOT00000024323
|
Rnf182
|
ring finger protein 182 |
| chr4_-_123713319 | 0.07 |
ENSRNOT00000012875
ENSRNOT00000086982 |
Slc6a6
|
solute carrier family 6 member 6 |
| chr20_+_32509598 | 0.07 |
ENSRNOT00000046268
|
Kpna5
|
karyopherin subunit alpha 5 |
| chr1_+_213886775 | 0.07 |
ENSRNOT00000020864
|
Pkp3
|
plakophilin 3 |
| chr12_-_29743705 | 0.06 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr8_+_99880073 | 0.06 |
ENSRNOT00000010765
|
Plscr4
|
phospholipid scramblase 4 |
| chr8_-_6305033 | 0.06 |
ENSRNOT00000029887
|
Cep126
|
centrosomal protein 126 |
| chr5_+_59080765 | 0.06 |
ENSRNOT00000021888
ENSRNOT00000064419 |
Rgp1
|
RGP1 homolog, RAB6A GEF complex partner 1 |
| chr4_-_177196180 | 0.05 |
ENSRNOT00000018999
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr7_+_74350479 | 0.05 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr18_+_29386809 | 0.05 |
ENSRNOT00000082079
ENSRNOT00000024825 |
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr15_-_61648267 | 0.05 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr19_+_38422164 | 0.05 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr4_+_70252366 | 0.05 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chr10_-_87407634 | 0.04 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr6_+_73358112 | 0.04 |
ENSRNOT00000041373
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr12_-_41590244 | 0.04 |
ENSRNOT00000038736
|
Slc8b1
|
solute carrier family 8 member B1 |
| chr20_-_3372070 | 0.04 |
ENSRNOT00000080770
|
Dhx16
|
DEAH-box helicase 16 |
| chr3_-_148057523 | 0.04 |
ENSRNOT00000055408
|
Defb24
|
defensin beta 24 |
| chr9_+_37462693 | 0.04 |
ENSRNOT00000016316
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr1_+_62133816 | 0.04 |
ENSRNOT00000034746
|
LOC102555377
|
zinc finger protein 182-like |
| chr8_+_69753373 | 0.04 |
ENSRNOT00000066973
|
Tipin
|
timeless interacting protein |
| chr7_-_134495835 | 0.04 |
ENSRNOT00000058349
|
Gxylt1
|
glucoside xylosyltransferase 1 |
| chr7_+_20262680 | 0.03 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr8_+_5967463 | 0.03 |
ENSRNOT00000086251
|
Tmem123
|
transmembrane protein 123 |
| chr1_-_211196868 | 0.03 |
ENSRNOT00000022714
|
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr15_+_1054937 | 0.03 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr16_-_73827488 | 0.03 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr8_-_76940094 | 0.03 |
ENSRNOT00000082709
ENSRNOT00000084313 |
Rnf111
|
ring finger protein 111 |
| chr10_-_73629581 | 0.03 |
ENSRNOT00000091172
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1 |
| chr16_-_81693000 | 0.02 |
ENSRNOT00000092353
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr5_+_135351816 | 0.02 |
ENSRNOT00000082602
|
Ipp
|
intracisternal A particle-promoted polypeptide |
| chrX_+_70438617 | 0.02 |
ENSRNOT00000076671
|
Arr3
|
arrestin 3 |
| chr15_+_39779648 | 0.02 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr1_+_87790104 | 0.02 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chr5_-_160620334 | 0.02 |
ENSRNOT00000019171
|
Tmem51
|
transmembrane protein 51 |
| chr2_-_112802073 | 0.02 |
ENSRNOT00000093081
|
Ect2
|
epithelial cell transforming 2 |
| chr6_-_98566523 | 0.02 |
ENSRNOT00000077715
|
Ppp2r5e
|
protein phosphatase 2 regulatory subunit B', epsilon |
| chr14_+_48726045 | 0.02 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_+_23012070 | 0.02 |
ENSRNOT00000042460
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr6_-_107080524 | 0.02 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr9_+_30939555 | 0.02 |
ENSRNOT00000016887
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr12_-_5685448 | 0.02 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chr5_-_100970043 | 0.02 |
ENSRNOT00000013950
|
Zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr5_+_130286393 | 0.02 |
ENSRNOT00000044795
|
Ccp6l1
|
cytosolic carboxypeptidase 6-like 1 |
| chr16_+_74534130 | 0.02 |
ENSRNOT00000050637
|
Slc25a15
|
solute carrier family 25 member 15 |
| chr19_-_37952501 | 0.02 |
ENSRNOT00000026809
|
Dpep3
|
dipeptidase 3 |
| chr10_+_108132105 | 0.02 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr6_+_126766967 | 0.02 |
ENSRNOT00000052296
|
Cox8c
|
cytochrome c oxidase subunit 8C |
| chr8_+_113939886 | 0.02 |
ENSRNOT00000079562
|
Aste1
|
asteroid homolog 1 (Drosophila) |
| chr12_-_44999074 | 0.02 |
ENSRNOT00000079177
ENSRNOT00000038293 |
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr14_-_78902063 | 0.02 |
ENSRNOT00000088469
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr19_-_52424318 | 0.01 |
ENSRNOT00000021743
|
Tldc1
|
TBC/LysM-associated domain containing 1 |
| chr20_-_5110191 | 0.01 |
ENSRNOT00000076597
|
Csnk2b
|
casein kinase 2 beta |
| chr15_-_61772516 | 0.01 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr6_+_8284878 | 0.01 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr3_-_153321352 | 0.01 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chrX_+_128550178 | 0.01 |
ENSRNOT00000082970
|
Stag2
|
stromal antigen 2 |
| chr3_+_54253949 | 0.01 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr6_-_42616548 | 0.01 |
ENSRNOT00000081433
|
Atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr1_+_30681681 | 0.01 |
ENSRNOT00000015395
|
Rspo3
|
R-spondin 3 |
| chr7_-_27552078 | 0.01 |
ENSRNOT00000059538
|
Stab2
|
stabilin 2 |
| chr9_+_95161157 | 0.01 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr13_-_78521911 | 0.01 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr8_-_116855062 | 0.01 |
ENSRNOT00000050344
|
Rnf123
|
ring finger protein 123 |
| chr8_-_53362006 | 0.01 |
ENSRNOT00000077145
|
LOC100360390
|
claudin 25-like |
| chr15_+_70790125 | 0.01 |
ENSRNOT00000012440
|
Tdrd3
|
tudor domain containing 3 |
| chr3_-_162581610 | 0.01 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr2_+_165601007 | 0.01 |
ENSRNOT00000013931
|
Smc4
|
structural maintenance of chromosomes 4 |
| chrX_-_14220662 | 0.01 |
ENSRNOT00000045753
|
Srpx
|
sushi-repeat-containing protein, X-linked |
| chr10_+_109665682 | 0.01 |
ENSRNOT00000054963
|
Slc25a10
|
solute carrier family 25 member 10 |
| chr7_-_140556639 | 0.01 |
ENSRNOT00000089523
|
Rhebl1
|
Ras homolog enriched in brain like 1 |
| chrX_+_43626480 | 0.01 |
ENSRNOT00000068078
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr10_+_83081168 | 0.01 |
ENSRNOT00000035023
|
Tac4
|
tachykinin 4 (hemokinin) |
| chr8_+_132120452 | 0.01 |
ENSRNOT00000055822
|
Tgm4
|
transglutaminase 4 |
| chr5_+_64566804 | 0.01 |
ENSRNOT00000073192
|
LOC103690035
|
TGF-beta receptor type-1-like |
| chr7_-_97071968 | 0.01 |
ENSRNOT00000006596
|
Slc22a22
|
solute carrier family 22 (organic cation transporter), member 22 |
| chr3_-_2490392 | 0.00 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr14_+_81819799 | 0.00 |
ENSRNOT00000076840
|
Mxd4
|
Max dimerization protein 4 |
| chr7_-_68549763 | 0.00 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr12_+_37702404 | 0.00 |
ENSRNOT00000049985
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr2_+_68820615 | 0.00 |
ENSRNOT00000087007
ENSRNOT00000089504 |
Egf
|
epidermal growth factor |
| chr1_+_68239314 | 0.00 |
ENSRNOT00000070823
|
Vom1r109
|
vomeronasal 1 receptor 109 |
| chr7_+_141237768 | 0.00 |
ENSRNOT00000091717
|
Aqp2
|
aquaporin 2 |
| chr2_+_68821004 | 0.00 |
ENSRNOT00000083713
|
Egf
|
epidermal growth factor |
| chr3_+_11207542 | 0.00 |
ENSRNOT00000013546
|
Prrc2b
|
proline-rich coiled-coil 2B |
| chrX_-_15651332 | 0.00 |
ENSRNOT00000039000
|
Gpkow
|
G patch domain and KOW motifs |
| chr8_+_22423467 | 0.00 |
ENSRNOT00000085320
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr6_-_105097054 | 0.00 |
ENSRNOT00000048606
|
Slc8a3
|
solute carrier family 8 member A3 |
| chr19_-_59894937 | 0.00 |
ENSRNOT00000027145
|
Rbm34
|
RNA binding motif protein 34 |
| chr16_+_54765325 | 0.00 |
ENSRNOT00000065327
ENSRNOT00000086899 |
Mtmr7
|
myotubularin related protein 7 |
| chr5_-_146795866 | 0.00 |
ENSRNOT00000065640
|
Tlr12
|
toll-like receptor 12 |
| chr3_+_140024043 | 0.00 |
ENSRNOT00000086409
|
Rin2
|
Ras and Rab interactor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfat5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030977 | taurine binding(GO:0030977) |