Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
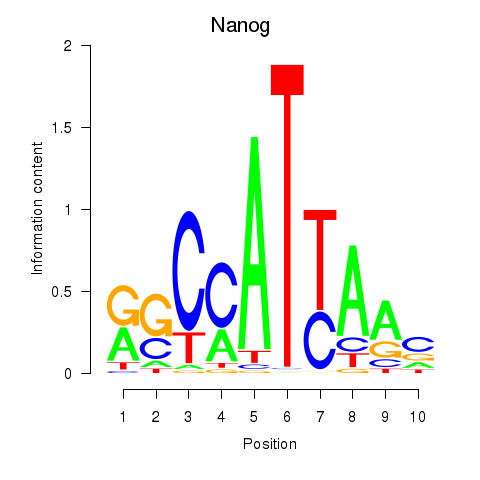
Results for Nanog
Z-value: 0.31
Transcription factors associated with Nanog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nanog
|
ENSRNOG00000008368 | Nanog homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nanog | rn6_v1_chr4_+_155531906_155531906 | 0.34 | 5.7e-01 | Click! |
Activity profile of Nanog motif
Sorted Z-values of Nanog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_123106694 | 0.26 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr4_+_171250818 | 0.14 |
ENSRNOT00000040576
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_87066289 | 0.10 |
ENSRNOT00000027645
|
Capn12
|
calpain 12 |
| chr8_+_72743426 | 0.09 |
ENSRNOT00000072573
|
Rps27l
|
ribosomal protein S27-like |
| chr2_+_187447501 | 0.09 |
ENSRNOT00000038589
|
Iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr1_-_168972725 | 0.09 |
ENSRNOT00000090422
|
Hbb
|
hemoglobin subunit beta |
| chr4_-_178168690 | 0.08 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr3_+_44025300 | 0.08 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr2_-_138833933 | 0.08 |
ENSRNOT00000013343
|
Pcdh18
|
protocadherin 18 |
| chrX_-_138148967 | 0.08 |
ENSRNOT00000033968
|
Frmd7
|
FERM domain containing 7 |
| chr7_-_143497108 | 0.08 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr5_+_144031402 | 0.07 |
ENSRNOT00000011694
|
Csf3r
|
colony stimulating factor 3 receptor |
| chr7_-_28715224 | 0.07 |
ENSRNOT00000065899
|
Parpbp
|
PARP1 binding protein |
| chr19_+_14345993 | 0.07 |
ENSRNOT00000084913
|
Gm18025
|
predicted gene, 18025 |
| chr9_+_20241062 | 0.07 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr5_-_136098013 | 0.07 |
ENSRNOT00000089166
|
RGD1563714
|
RGD1563714 |
| chr9_+_81644355 | 0.06 |
ENSRNOT00000071700
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr17_-_51912496 | 0.06 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr2_+_208750356 | 0.06 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr18_+_81694808 | 0.06 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr15_+_12827707 | 0.06 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr8_+_39687269 | 0.06 |
ENSRNOT00000012060
|
Tmem218
|
transmembrane protein 218 |
| chr1_+_91363492 | 0.06 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr3_+_150761329 | 0.05 |
ENSRNOT00000067854
|
Dynlrb1
|
dynein light chain roadblock-type 1 |
| chr4_-_170912629 | 0.05 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr2_+_54466280 | 0.05 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr3_+_100768637 | 0.05 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr10_-_15098791 | 0.05 |
ENSRNOT00000026139
|
Chtf18
|
chromosome transmission fidelity factor 18 |
| chr1_-_216663720 | 0.05 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr7_+_77066955 | 0.05 |
ENSRNOT00000008700
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr17_-_78735324 | 0.05 |
ENSRNOT00000036299
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr7_-_54855557 | 0.05 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr19_-_10450186 | 0.05 |
ENSRNOT00000088048
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chrX_+_13441558 | 0.05 |
ENSRNOT00000030170
|
RGD1561661
|
similar to Ferritin light chain (Ferritin L subunit) |
| chr3_-_161294125 | 0.05 |
ENSRNOT00000021143
|
Spata25
|
spermatogenesis associated 25 |
| chr6_-_47904163 | 0.05 |
ENSRNOT00000011333
|
Rps7
|
ribosomal protein S7 |
| chr10_-_89005213 | 0.05 |
ENSRNOT00000027159
|
Psmc3ip
|
PSMC3 interacting protein |
| chr17_-_42031265 | 0.05 |
ENSRNOT00000068021
|
Dcdc2
|
doublecortin domain containing 2 |
| chr10_+_89635675 | 0.05 |
ENSRNOT00000028179
|
RGD1561590
|
similar to SAP18 |
| chr3_+_138174054 | 0.04 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr7_+_140054592 | 0.04 |
ENSRNOT00000014337
|
Olr1108
|
olfactory receptor 1108 |
| chrX_-_142164220 | 0.04 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr2_-_219262901 | 0.04 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chrM_+_5323 | 0.04 |
ENSRNOT00000050156
|
Mt-co1
|
mitochondrially encoded cytochrome c oxidase 1 |
| chr10_+_59566223 | 0.04 |
ENSRNOT00000024068
ENSRNOT00000088245 |
P2rx1
|
purinergic receptor P2X 1 |
| chr20_+_27592379 | 0.04 |
ENSRNOT00000047000
|
Trappc3l
|
trafficking protein particle complex 3-like |
| chr7_-_143228060 | 0.04 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr16_+_2706428 | 0.04 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chrX_+_123751293 | 0.04 |
ENSRNOT00000089883
|
Nkap
|
NFKB activating protein |
| chrX_-_115496045 | 0.04 |
ENSRNOT00000051134
|
LOC100361854
|
ribosomal protein S26-like |
| chr7_+_112057030 | 0.04 |
ENSRNOT00000035216
|
AABR07058366.1
|
|
| chr9_-_55673704 | 0.04 |
ENSRNOT00000066231
ENSRNOT00000081677 |
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2 |
| chr6_+_132551394 | 0.04 |
ENSRNOT00000019583
|
Evl
|
Enah/Vasp-like |
| chr2_-_244370983 | 0.04 |
ENSRNOT00000021458
|
Rap1gds1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr10_-_87541851 | 0.04 |
ENSRNOT00000089610
|
RGD1561684
|
similar to keratin associated protein 2-4 |
| chrX_-_106607352 | 0.04 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chrX_+_105147534 | 0.04 |
ENSRNOT00000046288
|
Cenpi
|
centromere protein I |
| chr10_+_105498728 | 0.04 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr20_+_5374985 | 0.04 |
ENSRNOT00000052270
|
RT1-A2
|
RT1 class Ia, locus A2 |
| chr1_+_82473737 | 0.04 |
ENSRNOT00000028029
|
B9d2
|
B9 protein domain 2 |
| chr7_+_24939498 | 0.04 |
ENSRNOT00000010601
|
Ckap4
|
cytoskeleton-associated protein 4 |
| chr10_+_45297937 | 0.04 |
ENSRNOT00000066367
|
Trim17
|
tripartite motif-containing 17 |
| chr9_-_82336806 | 0.04 |
ENSRNOT00000024667
|
Slc23a3
|
solute carrier family 23, member 3 |
| chr14_-_21709084 | 0.04 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chrX_+_123751089 | 0.04 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr2_+_189609800 | 0.03 |
ENSRNOT00000089016
|
Slc39a1
|
solute carrier family 39 member 1 |
| chr8_-_115145810 | 0.03 |
ENSRNOT00000079470
|
Abhd14a
|
abhydrolase domain containing 14A |
| chr1_-_178367828 | 0.03 |
ENSRNOT00000050823
|
AABR07071968.1
|
|
| chr2_+_208749996 | 0.03 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr1_-_255557055 | 0.03 |
ENSRNOT00000033780
|
Fgfbp3
|
fibroblast growth factor binding protein 3 |
| chr5_+_139038134 | 0.03 |
ENSRNOT00000078155
|
Hivep3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr5_-_115387377 | 0.03 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr7_+_34326087 | 0.03 |
ENSRNOT00000006971
|
Hal
|
histidine ammonia lyase |
| chr4_-_155401480 | 0.03 |
ENSRNOT00000020735
|
Apobec1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr1_+_247869756 | 0.03 |
ENSRNOT00000054764
|
Mlana
|
melan-A |
| chr14_+_23480462 | 0.03 |
ENSRNOT00000002755
|
Gnrhr
|
gonadotropin releasing hormone receptor |
| chr1_+_170147300 | 0.03 |
ENSRNOT00000048075
|
Olr200
|
olfactory receptor 200 |
| chr2_-_250778269 | 0.03 |
ENSRNOT00000085670
ENSRNOT00000083535 ENSRNOT00000017824 |
Clca5
|
chloride channel accessory 5 |
| chr7_+_29435444 | 0.03 |
ENSRNOT00000008613
|
Slc5a8
|
solute carrier family 5 member 8 |
| chr9_+_61552374 | 0.03 |
ENSRNOT00000050090
|
Tmem258b
|
transmembrane protein 258 B |
| chr4_+_110699557 | 0.03 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr10_+_34402482 | 0.03 |
ENSRNOT00000072317
|
Tpcr12
|
putative olfactory receptor |
| chr14_-_104612597 | 0.03 |
ENSRNOT00000007002
|
Slc1a4
|
solute carrier family 1 member 4 |
| chr3_+_67849966 | 0.03 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr4_+_163174487 | 0.03 |
ENSRNOT00000088108
|
Clec9a
|
C-type lectin domain family 9, member A |
| chr20_+_26999795 | 0.03 |
ENSRNOT00000057872
|
Mypn
|
myopalladin |
| chr8_+_48699769 | 0.03 |
ENSRNOT00000044613
|
Hyou1
|
hypoxia up-regulated 1 |
| chr7_-_136853957 | 0.03 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr9_+_12740885 | 0.03 |
ENSRNOT00000015210
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr19_-_10212050 | 0.03 |
ENSRNOT00000082822
|
Tepp
|
testis, prostate and placenta expressed |
| chr6_+_8669722 | 0.03 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr9_+_27068443 | 0.03 |
ENSRNOT00000017393
|
Efhc1
|
EF-hand domain containing 1 |
| chr5_-_155709215 | 0.03 |
ENSRNOT00000018118
|
Cdc42
|
cell division cycle 42 |
| chr2_+_216863428 | 0.03 |
ENSRNOT00000068413
|
Col11a1
|
collagen type XI alpha 1 chain |
| chr9_+_74124016 | 0.03 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr6_+_22696397 | 0.03 |
ENSRNOT00000011630
|
Alk
|
anaplastic lymphoma receptor tyrosine kinase |
| chr3_+_72080630 | 0.03 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chrX_+_136466779 | 0.03 |
ENSRNOT00000093268
ENSRNOT00000068717 |
Arhgap36
|
Rho GTPase activating protein 36 |
| chr2_+_60461551 | 0.03 |
ENSRNOT00000024557
|
Rad1
|
RAD1 checkpoint DNA exonuclease |
| chr3_-_23020441 | 0.03 |
ENSRNOT00000017651
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr2_-_112831476 | 0.03 |
ENSRNOT00000018055
|
Ect2
|
epithelial cell transforming 2 |
| chr8_-_116391158 | 0.03 |
ENSRNOT00000078720
ENSRNOT00000022550 |
Gnai2
|
G protein subunit alpha i2 |
| chr5_+_172364421 | 0.03 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr3_+_147511563 | 0.03 |
ENSRNOT00000006828
|
Slc52a3
|
solute carrier family 52 member 3 |
| chr20_+_4178686 | 0.03 |
ENSRNOT00000080641
|
Tesb
|
testis specific basic protein |
| chr19_-_27535792 | 0.03 |
ENSRNOT00000024040
|
Olr1666
|
olfactory receptor 1666 |
| chr1_-_1889236 | 0.03 |
ENSRNOT00000087944
|
AABR07000159.2
|
|
| chr19_-_28645488 | 0.03 |
ENSRNOT00000044160
|
AABR07043449.1
|
|
| chr9_+_43259709 | 0.03 |
ENSRNOT00000022487
|
Cox5b
|
cytochrome c oxidase subunit 5B |
| chr3_+_114918648 | 0.03 |
ENSRNOT00000089204
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr7_-_60294796 | 0.03 |
ENSRNOT00000007580
|
Yeats4
|
YEATS domain containing 4 |
| chrX_-_123708211 | 0.03 |
ENSRNOT00000092456
|
Rpl39
|
ribosomal protein L39 |
| chr3_-_127500709 | 0.03 |
ENSRNOT00000006330
|
Hao1
|
hydroxyacid oxidase 1 |
| chr1_+_215610368 | 0.03 |
ENSRNOT00000078903
ENSRNOT00000087781 |
Tnni2
|
troponin I2, fast skeletal type |
| chr1_-_64021321 | 0.03 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr1_-_100231460 | 0.03 |
ENSRNOT00000032200
|
Acpt
|
acid phosphatase, testicular |
| chr1_+_64162672 | 0.03 |
ENSRNOT00000089713
|
Tfpt
|
TCF3 (E2A) fusion partner |
| chr5_+_98387291 | 0.03 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr13_+_47935163 | 0.03 |
ENSRNOT00000006768
|
Eif2d
|
eukaryotic translation initiation factor 2D |
| chr8_-_128711221 | 0.03 |
ENSRNOT00000055888
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr19_-_25378974 | 0.03 |
ENSRNOT00000011150
|
Ccdc130
|
coiled-coil domain containing 130 |
| chr8_+_55504857 | 0.03 |
ENSRNOT00000088320
|
Btg4
|
BTG anti-proliferation factor 4 |
| chr3_-_46601409 | 0.03 |
ENSRNOT00000079261
|
Pla2r1
|
phospholipase A2 receptor 1 |
| chr8_+_55037750 | 0.03 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chrX_+_77065397 | 0.03 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr2_-_118882562 | 0.02 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr5_-_154332940 | 0.02 |
ENSRNOT00000014458
|
Pithd1
|
PITH domain containing 1 |
| chr8_-_72204730 | 0.02 |
ENSRNOT00000023810
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr9_-_23493081 | 0.02 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr10_+_45966926 | 0.02 |
ENSRNOT00000067584
|
Olfr225
|
olfactory receptor 225 |
| chr5_-_141430659 | 0.02 |
ENSRNOT00000034944
|
Akirin1
|
akirin 1 |
| chr10_-_34333305 | 0.02 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chr4_-_34282351 | 0.02 |
ENSRNOT00000011123
|
Rpa3
|
replication protein A3 |
| chrM_+_9451 | 0.02 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr5_-_24631679 | 0.02 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr15_-_52320385 | 0.02 |
ENSRNOT00000067776
|
Dmtn
|
dematin actin binding protein |
| chr8_-_132919110 | 0.02 |
ENSRNOT00000008747
|
Xcr1
|
X-C motif chemokine receptor 1 |
| chr1_+_101214593 | 0.02 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr17_-_62299655 | 0.02 |
ENSRNOT00000024855
|
Gjd4
|
gap junction protein, delta 4 |
| chr8_+_117455262 | 0.02 |
ENSRNOT00000027520
|
Slc25a20
|
solute carrier family 25 member 20 |
| chr1_+_222844144 | 0.02 |
ENSRNOT00000032879
|
Pla2g16
|
phospholipase A2, group XVI |
| chr10_+_43851950 | 0.02 |
ENSRNOT00000085666
|
LOC691277
|
similar to Robo-1 |
| chr4_+_153805993 | 0.02 |
ENSRNOT00000056174
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr5_-_101166651 | 0.02 |
ENSRNOT00000078862
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr10_-_90151042 | 0.02 |
ENSRNOT00000055187
|
Hdac5
|
histone deacetylase 5 |
| chr1_-_19376301 | 0.02 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr11_-_32550539 | 0.02 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr1_+_177569618 | 0.02 |
ENSRNOT00000090042
|
Tead1
|
TEA domain transcription factor 1 |
| chr5_+_59008933 | 0.02 |
ENSRNOT00000023060
|
Car9
|
carbonic anhydrase 9 |
| chr13_-_90977734 | 0.02 |
ENSRNOT00000011869
|
Slamf8
|
SLAM family member 8 |
| chr16_+_71738718 | 0.02 |
ENSRNOT00000022097
|
Plekha2
|
pleckstrin homology domain containing A2 |
| chr14_-_19072677 | 0.02 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr1_-_89014189 | 0.02 |
ENSRNOT00000028413
|
Lin37
|
lin-37 DREAM MuvB core complex component |
| chr9_-_99299715 | 0.02 |
ENSRNOT00000027622
|
Hdac4
|
histone deacetylase 4 |
| chr11_+_87549059 | 0.02 |
ENSRNOT00000002567
|
Ccdc74a
|
coiled-coil domain containing 74A |
| chr10_+_68588789 | 0.02 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr2_-_195279218 | 0.02 |
ENSRNOT00000085565
|
RGD1559714
|
similar to TDPOZ3 |
| chr5_+_33097654 | 0.02 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr13_-_48927483 | 0.02 |
ENSRNOT00000010976
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr19_-_29134876 | 0.02 |
ENSRNOT00000040545
|
AABR07043523.1
|
|
| chr3_-_147263275 | 0.02 |
ENSRNOT00000029620
|
Psmf1
|
proteasome inhibitor subunit 1 |
| chr7_+_78649875 | 0.02 |
ENSRNOT00000006243
|
Dcstamp
|
dendrocyte expressed seven transmembrane protein |
| chr7_-_134722215 | 0.02 |
ENSRNOT00000036750
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chrX_-_1346181 | 0.02 |
ENSRNOT00000042736
|
LOC102555453
|
60S ribosomal protein L12-like |
| chr4_-_170740274 | 0.02 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr10_+_49231730 | 0.02 |
ENSRNOT00000065335
|
Trim16
|
tripartite motif-containing 16 |
| chr1_+_60717386 | 0.02 |
ENSRNOT00000015019
|
Ppp2r1a
|
protein phosphatase 2 scaffold subunit A alpha |
| chr10_+_62303662 | 0.02 |
ENSRNOT00000056355
|
Tlcd2
|
TLC domain containing 2 |
| chr8_-_49502647 | 0.02 |
ENSRNOT00000040313
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr11_+_76147205 | 0.02 |
ENSRNOT00000071586
|
Fgf12
|
fibroblast growth factor 12 |
| chr1_-_100234040 | 0.02 |
ENSRNOT00000079199
|
Acpt
|
acid phosphatase, testicular |
| chr5_+_33784715 | 0.02 |
ENSRNOT00000035685
|
Slc7a13
|
solute carrier family 7 member 13 |
| chrX_-_23139694 | 0.02 |
ENSRNOT00000033656
|
Pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr16_-_47535358 | 0.02 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr4_-_138830117 | 0.02 |
ENSRNOT00000008403
|
Il5ra
|
interleukin 5 receptor subunit alpha |
| chr14_+_45062662 | 0.02 |
ENSRNOT00000059124
|
Tlr10
|
toll-like receptor 10 |
| chr20_+_26534535 | 0.02 |
ENSRNOT00000000423
|
Tmem167b
|
transmembrane protein 167B |
| chr3_-_147481073 | 0.02 |
ENSRNOT00000084722
|
Fam110a
|
family with sequence similarity 110, member A |
| chr10_+_31324512 | 0.02 |
ENSRNOT00000008559
|
Fndc9
|
fibronectin type III domain containing 9 |
| chr5_+_154668179 | 0.02 |
ENSRNOT00000015971
|
Hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr4_+_118167294 | 0.02 |
ENSRNOT00000022367
|
LOC687679
|
similar to small nuclear ribonucleoprotein polypeptide G |
| chr2_-_179704629 | 0.02 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr5_-_151768123 | 0.02 |
ENSRNOT00000079380
|
Nudc
|
nuclear distribution C, dynein complex regulator |
| chr19_+_54314865 | 0.02 |
ENSRNOT00000024069
|
Irf8
|
interferon regulatory factor 8 |
| chr19_-_14945302 | 0.02 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr7_+_60015998 | 0.02 |
ENSRNOT00000007309
|
Best3
|
bestrophin 3 |
| chr17_+_2690062 | 0.02 |
ENSRNOT00000024937
|
Olr1653
|
olfactory receptor 1653 |
| chr8_-_48672732 | 0.02 |
ENSRNOT00000079275
|
Hmbs
|
hydroxymethylbilane synthase |
| chr7_-_58343137 | 0.02 |
ENSRNOT00000005310
ENSRNOT00000090560 |
Tmem19
|
transmembrane protein 19 |
| chr16_-_31972507 | 0.02 |
ENSRNOT00000032199
|
Cbr4
|
carbonyl reductase 4 |
| chr18_-_69944632 | 0.02 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr15_-_9086282 | 0.02 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr13_+_99335020 | 0.02 |
ENSRNOT00000029787
|
AABR07021930.1
|
|
| chrX_-_152932953 | 0.02 |
ENSRNOT00000078929
|
Cetn2
|
centrin 2 |
| chr8_+_115069095 | 0.02 |
ENSRNOT00000014770
|
Dusp7
|
dual specificity phosphatase 7 |
| chr3_+_117421604 | 0.02 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr20_-_157665 | 0.02 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chr1_-_214511529 | 0.02 |
ENSRNOT00000026390
|
Chid1
|
chitinase domain containing 1 |
| chr2_+_178679041 | 0.02 |
ENSRNOT00000013524
|
Fam198b
|
family with sequence similarity 198, member B |
| chr1_+_189432604 | 0.02 |
ENSRNOT00000034610
|
Acsm4
|
acyl-CoA synthetase medium-chain family member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nanog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.1 | GO:0010999 | regulation of eIF2 alpha phosphorylation by heme(GO:0010999) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:2000741 | regulation of timing of neuron differentiation(GO:0060164) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:0002554 | serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.0 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) formamide metabolic process(GO:0043606) |
| 0.0 | 0.0 | GO:0060661 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.0 | GO:0090648 | response to environmental enrichment(GO:0090648) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.0 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |