Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
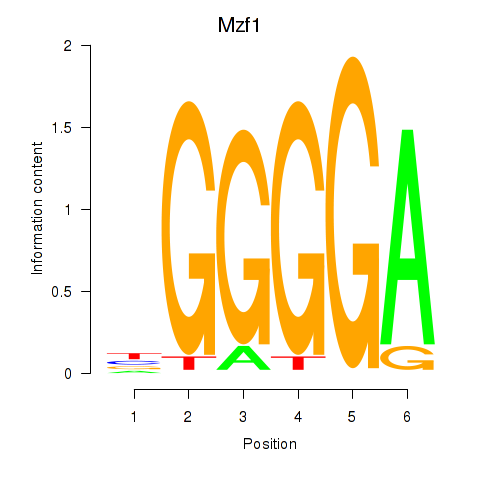
Results for Mzf1
Z-value: 0.80
Transcription factors associated with Mzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mzf1
|
ENSRNOG00000027550 | myeloid zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mzf1 | rn6_v1_chr1_+_65522118_65522118 | 0.28 | 6.5e-01 | Click! |
Activity profile of Mzf1 motif
Sorted Z-values of Mzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_109300433 | 0.44 |
ENSRNOT00000010712
|
Fos
|
FBJ osteosarcoma oncogene |
| chr10_-_85574889 | 0.42 |
ENSRNOT00000072274
|
LOC691153
|
hypothetical protein LOC691153 |
| chr7_+_70364813 | 0.40 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr2_+_210381829 | 0.39 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chr7_+_142912316 | 0.36 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr9_+_17841410 | 0.26 |
ENSRNOT00000031706
|
Tmem151b
|
transmembrane protein 151B |
| chr2_+_196334626 | 0.25 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chrX_+_76786466 | 0.23 |
ENSRNOT00000090665
|
Fgf16
|
fibroblast growth factor 16 |
| chr2_-_77632628 | 0.22 |
ENSRNOT00000073915
|
Basp1
|
brain abundant, membrane attached signal protein 1 |
| chr19_+_59328960 | 0.22 |
ENSRNOT00000086865
|
Coa6
|
cytochrome c oxidase assembly factor 6 |
| chr10_-_64657089 | 0.20 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr16_-_14300951 | 0.20 |
ENSRNOT00000017038
|
Rgr
|
retinal G protein coupled receptor |
| chr20_+_4993560 | 0.19 |
ENSRNOT00000081628
ENSRNOT00000087861 ENSRNOT00000001160 |
Vars
|
valyl-tRNA synthetase |
| chr7_-_143967484 | 0.19 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr10_+_85301875 | 0.19 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr5_+_58393603 | 0.18 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr15_-_33725188 | 0.18 |
ENSRNOT00000083941
|
Zfhx2
|
zinc finger homeobox 2 |
| chr6_+_9483594 | 0.17 |
ENSRNOT00000089272
|
AABR07062799.2
|
|
| chr8_+_48805684 | 0.15 |
ENSRNOT00000064041
|
Bcl9l
|
B-cell CLL/lymphoma 9-like |
| chr19_+_23389375 | 0.15 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr3_+_170550314 | 0.15 |
ENSRNOT00000006991
|
Tfap2c
|
transcription factor AP-2 gamma |
| chr1_+_101783621 | 0.14 |
ENSRNOT00000067679
|
Lmtk3
|
lemur tyrosine kinase 3 |
| chr5_-_59085676 | 0.14 |
ENSRNOT00000068105
|
Msmp
|
microseminoprotein, prostate associated |
| chr17_-_90627101 | 0.13 |
ENSRNOT00000003349
|
Nid1
|
nidogen 1 |
| chr4_+_142453013 | 0.13 |
ENSRNOT00000056573
|
AABR07061755.1
|
|
| chr18_+_30890869 | 0.13 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr10_+_56627411 | 0.13 |
ENSRNOT00000089108
ENSRNOT00000068493 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr1_-_101388125 | 0.13 |
ENSRNOT00000028176
ENSRNOT00000049957 |
Snrnp70
|
small nuclear ribonucleoprotein U1 subunit 70 |
| chr5_+_144160108 | 0.13 |
ENSRNOT00000064972
|
Eva1b
|
eva-1 homolog B |
| chr10_-_56444847 | 0.12 |
ENSRNOT00000056872
ENSRNOT00000092662 |
Nlgn2
|
neuroligin 2 |
| chr3_-_7795758 | 0.12 |
ENSRNOT00000045919
|
Ntng2
|
netrin G2 |
| chr2_-_188559882 | 0.12 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr1_-_48360131 | 0.12 |
ENSRNOT00000051927
ENSRNOT00000023116 |
Slc22a2
|
solute carrier family 22 member 2 |
| chr8_-_63291966 | 0.11 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr19_+_54060622 | 0.11 |
ENSRNOT00000023607
|
Gse1
|
Gse1 coiled-coil protein |
| chr2_+_206064179 | 0.11 |
ENSRNOT00000025953
|
Syt6
|
synaptotagmin 6 |
| chr15_+_23665202 | 0.11 |
ENSRNOT00000089226
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr2_-_186245342 | 0.11 |
ENSRNOT00000057062
ENSRNOT00000022292 |
Dclk2
|
doublecortin-like kinase 2 |
| chr1_+_80000165 | 0.10 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr6_-_21600451 | 0.10 |
ENSRNOT00000047674
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_10307265 | 0.10 |
ENSRNOT00000092627
|
Wasf3
|
WAS protein family, member 3 |
| chr14_-_35652709 | 0.10 |
ENSRNOT00000003080
|
Gsx2
|
GS homeobox 2 |
| chr7_-_121232741 | 0.10 |
ENSRNOT00000023196
|
Pdgfb
|
platelet derived growth factor subunit B |
| chr10_-_91047177 | 0.10 |
ENSRNOT00000003986
|
C1ql1
|
complement C1q like 1 |
| chr7_-_3074359 | 0.10 |
ENSRNOT00000019738
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr7_-_36499784 | 0.09 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr7_+_142869752 | 0.09 |
ENSRNOT00000009979
|
Grasp
|
general receptor for phosphoinositides 1 associated scaffold protein |
| chr10_+_97212432 | 0.09 |
ENSRNOT00000088599
|
Axin2
|
axin 2 |
| chr10_-_85684138 | 0.09 |
ENSRNOT00000017989
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase type 2 beta |
| chr2_-_186245163 | 0.09 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr20_+_5050327 | 0.09 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr10_-_110182291 | 0.09 |
ENSRNOT00000015178
ENSRNOT00000054936 |
Csnk1d
|
casein kinase 1, delta |
| chr8_+_45797315 | 0.09 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr1_-_72377434 | 0.09 |
ENSRNOT00000022193
|
Zfp524
|
zinc finger protein 524 |
| chrX_+_70596901 | 0.08 |
ENSRNOT00000088114
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr1_+_140998240 | 0.08 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chrX_-_157095274 | 0.08 |
ENSRNOT00000084088
|
Abcd1
|
ATP binding cassette subfamily D member 1 |
| chr2_+_206064394 | 0.08 |
ENSRNOT00000077739
|
Syt6
|
synaptotagmin 6 |
| chr12_-_19582185 | 0.08 |
ENSRNOT00000060020
ENSRNOT00000085853 |
RGD1305455
|
similar to hypothetical protein FLJ10925 |
| chr7_+_58366192 | 0.08 |
ENSRNOT00000081891
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr11_-_83926524 | 0.08 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr2_-_198719202 | 0.08 |
ENSRNOT00000028801
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr12_+_21721837 | 0.07 |
ENSRNOT00000073353
|
Mepce
|
methylphosphate capping enzyme |
| chr10_-_65963932 | 0.07 |
ENSRNOT00000011726
|
Nlk
|
nemo like kinase |
| chr10_-_85084850 | 0.07 |
ENSRNOT00000012462
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr7_-_140546908 | 0.07 |
ENSRNOT00000077502
|
Kmt2d
|
lysine methyltransferase 2D |
| chr2_-_207300854 | 0.07 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr1_+_44446765 | 0.07 |
ENSRNOT00000030132
|
Cldn20
|
claudin 20 |
| chr9_+_82033543 | 0.07 |
ENSRNOT00000023439
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr2_+_113984824 | 0.07 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr13_-_72367980 | 0.07 |
ENSRNOT00000003928
|
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr11_-_65209268 | 0.06 |
ENSRNOT00000077612
|
Gsk3b
|
glycogen synthase kinase 3 beta |
| chr2_+_188844073 | 0.06 |
ENSRNOT00000028117
|
Kcnn3
|
potassium calcium-activated channel subfamily N member 3 |
| chrX_-_134719097 | 0.06 |
ENSRNOT00000068478
|
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr8_-_111850075 | 0.06 |
ENSRNOT00000082097
|
Cdv3
|
CDV3 homolog |
| chr1_+_278198179 | 0.06 |
ENSRNOT00000023247
|
Fam160b1
|
family with sequence similarity 160, member B1 |
| chr18_+_47739140 | 0.06 |
ENSRNOT00000082760
|
Sncaip
|
synuclein, alpha interacting protein |
| chr1_+_82480195 | 0.06 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr5_-_146069670 | 0.06 |
ENSRNOT00000072793
|
LOC682102
|
hypothetical protein LOC682102 |
| chr10_+_85744568 | 0.06 |
ENSRNOT00000005522
|
Lasp1
|
LIM and SH3 protein 1 |
| chr5_+_147476221 | 0.06 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr7_+_23403891 | 0.05 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr9_-_10991820 | 0.05 |
ENSRNOT00000073011
ENSRNOT00000081441 |
Hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr4_+_56625561 | 0.05 |
ENSRNOT00000008356
ENSRNOT00000090808 ENSRNOT00000008972 |
Calu
|
calumenin |
| chr5_+_59080765 | 0.05 |
ENSRNOT00000021888
ENSRNOT00000064419 |
Rgp1
|
RGP1 homolog, RAB6A GEF complex partner 1 |
| chr4_-_153028129 | 0.05 |
ENSRNOT00000074558
|
Cecr6
|
cat eye syndrome chromosome region, candidate 6 |
| chr2_-_199354793 | 0.05 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr15_+_12827707 | 0.05 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr6_+_132702448 | 0.05 |
ENSRNOT00000005743
|
Yy1
|
YY1 transcription factor |
| chr11_+_57207656 | 0.05 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr1_+_144831523 | 0.05 |
ENSRNOT00000039748
|
Mex3b
|
mex-3 RNA binding family member B |
| chr17_-_90217786 | 0.05 |
ENSRNOT00000073534
|
Impad1
|
inositol monophosphatase domain containing 1 |
| chr15_-_52317219 | 0.05 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr10_+_86860685 | 0.04 |
ENSRNOT00000084644
|
Rara
|
retinoic acid receptor, alpha |
| chr2_-_46544457 | 0.04 |
ENSRNOT00000015680
|
Fst
|
follistatin |
| chr9_+_53627208 | 0.04 |
ENSRNOT00000083487
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr7_+_117759083 | 0.04 |
ENSRNOT00000050556
|
Gpt
|
glutamic--pyruvic transaminase |
| chr10_+_86157608 | 0.04 |
ENSRNOT00000082668
ENSRNOT00000008256 |
Cdk12
|
cyclin-dependent kinase 12 |
| chr5_-_136965191 | 0.04 |
ENSRNOT00000056842
|
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr3_-_57119001 | 0.04 |
ENSRNOT00000083171
|
Tlk1
|
tousled-like kinase 1 |
| chr20_-_1980101 | 0.04 |
ENSRNOT00000084582
ENSRNOT00000085050 ENSRNOT00000082545 ENSRNOT00000088396 |
Gabbr1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr10_+_66690133 | 0.04 |
ENSRNOT00000046262
|
Nf1
|
neurofibromin 1 |
| chr17_-_9762813 | 0.04 |
ENSRNOT00000033749
|
Slc34a1
|
solute carrier family 34 member 1 |
| chr1_+_15412603 | 0.03 |
ENSRNOT00000051496
ENSRNOT00000067070 |
Map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr10_-_56300077 | 0.03 |
ENSRNOT00000064401
|
Tnfsf12
|
tumor necrosis factor superfamily member 12 |
| chr6_-_26241337 | 0.03 |
ENSRNOT00000006525
|
Slc4a1ap
|
solute carrier family 4 member 1 adaptor protein |
| chr1_-_170175350 | 0.03 |
ENSRNOT00000023349
|
Olr201
|
olfactory receptor 201 |
| chr10_-_64862268 | 0.03 |
ENSRNOT00000056234
|
Phf12
|
PHD finger protein 12 |
| chr2_+_188748359 | 0.03 |
ENSRNOT00000028038
|
Shc1
|
SHC adaptor protein 1 |
| chr4_-_115157263 | 0.03 |
ENSRNOT00000015296
|
Tet3
|
tet methylcytosine dioxygenase 3 |
| chr7_-_70969905 | 0.03 |
ENSRNOT00000057745
|
Nab2
|
Ngfi-A binding protein 2 |
| chr9_+_69497121 | 0.03 |
ENSRNOT00000042562
|
Nrp2
|
neuropilin 2 |
| chr9_+_20264418 | 0.03 |
ENSRNOT00000051841
|
Atn1
|
atrophin 1 |
| chr10_-_88667345 | 0.03 |
ENSRNOT00000025518
|
Kcnh4
|
potassium voltage-gated channel subfamily H member 4 |
| chr10_-_11085142 | 0.03 |
ENSRNOT00000081596
ENSRNOT00000005479 ENSRNOT00000085230 |
Dnaja3
|
DnaJ heat shock protein family (Hsp40) member A3 |
| chr5_-_58113553 | 0.03 |
ENSRNOT00000046589
|
Arid3c
|
AT-rich interaction domain 3C |
| chr14_+_108007724 | 0.03 |
ENSRNOT00000014062
|
Xpo1
|
exportin 1 |
| chr4_+_145017608 | 0.03 |
ENSRNOT00000066723
|
Setd5
|
SET domain containing 5 |
| chr1_+_88182155 | 0.03 |
ENSRNOT00000083176
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr2_+_113984646 | 0.03 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr1_+_87180624 | 0.03 |
ENSRNOT00000056933
ENSRNOT00000027987 |
LOC103689986
|
protein YIF1B |
| chr1_-_72727112 | 0.03 |
ENSRNOT00000031172
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr6_-_105518725 | 0.03 |
ENSRNOT00000009693
ENSRNOT00000083488 |
Map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr10_+_46722109 | 0.03 |
ENSRNOT00000032685
|
Drc3
|
dynein regulatory complex subunit 3 |
| chr7_-_50638798 | 0.02 |
ENSRNOT00000048880
|
Syt1
|
synaptotagmin 1 |
| chr2_+_188495629 | 0.02 |
ENSRNOT00000027828
|
Fam189b
|
family with sequence similarity 189, member B |
| chr12_+_46615649 | 0.02 |
ENSRNOT00000086531
ENSRNOT00000089494 |
Bicdl1
|
BICD family like cargo adaptor 1 |
| chr9_+_62291809 | 0.02 |
ENSRNOT00000090621
|
Plcl1
|
phospholipase C-like 1 |
| chr15_+_33555640 | 0.02 |
ENSRNOT00000021096
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chrX_-_1856258 | 0.02 |
ENSRNOT00000079980
|
RGD1564855
|
similar to RuvB-like protein 1 |
| chr15_-_45927804 | 0.02 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr9_+_42620006 | 0.02 |
ENSRNOT00000019966
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr7_+_25919867 | 0.02 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr14_-_86868598 | 0.02 |
ENSRNOT00000087212
|
Nacad
|
NAC alpha domain containing |
| chr18_-_6474990 | 0.02 |
ENSRNOT00000061504
|
Kctd1
|
potassium channel tetramerization domain containing 1 |
| chr9_+_81518584 | 0.02 |
ENSRNOT00000084309
|
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr10_-_86144880 | 0.02 |
ENSRNOT00000067901
|
Med1
|
mediator complex subunit 1 |
| chr5_-_85123829 | 0.01 |
ENSRNOT00000007578
|
Brinp1
|
BMP/retinoic acid inducible neural specific 1 |
| chr20_+_5374985 | 0.01 |
ENSRNOT00000052270
|
RT1-A2
|
RT1 class Ia, locus A2 |
| chr3_-_156340913 | 0.01 |
ENSRNOT00000021452
|
Mafb
|
MAF bZIP transcription factor B |
| chr4_+_8256611 | 0.01 |
ENSRNOT00000061894
|
AABR07059198.1
|
|
| chr19_+_26142720 | 0.01 |
ENSRNOT00000005270
|
RGD1564093
|
similar to RIKEN cDNA 2310036O22 |
| chrX_+_70596576 | 0.01 |
ENSRNOT00000045082
ENSRNOT00000003741 ENSRNOT00000076079 |
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr1_+_199360645 | 0.01 |
ENSRNOT00000026527
|
Kat8
|
lysine acetyltransferase 8 |
| chr3_-_8659102 | 0.01 |
ENSRNOT00000050908
|
Zdhhc12
|
zinc finger, DHHC-type containing 12 |
| chr5_-_129113132 | 0.01 |
ENSRNOT00000080048
|
Rnf11l1
|
ring finger protein 11-like 1 |
| chr12_+_22229079 | 0.01 |
ENSRNOT00000001911
|
Gnb2
|
G protein subunit beta 2 |
| chr20_-_8574082 | 0.01 |
ENSRNOT00000048845
|
Mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr7_-_144880092 | 0.01 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr6_-_109692218 | 0.01 |
ENSRNOT00000012327
|
RGD1310769
|
similar to HSPC288 |
| chr20_-_5140304 | 0.01 |
ENSRNOT00000092646
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr7_-_77372398 | 0.01 |
ENSRNOT00000079644
ENSRNOT00000007999 |
Azin1
|
antizyme inhibitor 1 |
| chr20_+_7484550 | 0.01 |
ENSRNOT00000044395
|
Anks1a
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr4_-_83972540 | 0.01 |
ENSRNOT00000036951
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr3_-_119135391 | 0.01 |
ENSRNOT00000045443
|
Gabpb1
|
GA binding protein transcription factor, beta subunit 1 |
| chr14_+_4125380 | 0.01 |
ENSRNOT00000002882
|
Zfp644
|
zinc finger protein 644 |
| chr9_+_82700468 | 0.01 |
ENSRNOT00000027227
|
Inha
|
inhibin alpha subunit |
| chr1_+_183892841 | 0.00 |
ENSRNOT00000015498
|
Pde3b
|
phosphodiesterase 3B |
| chr16_-_82439441 | 0.00 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr8_+_52753011 | 0.00 |
ENSRNOT00000047264
|
Nxpe1
|
neurexophilin and PC-esterase domain family, member 1 |
| chr3_+_176691329 | 0.00 |
ENSRNOT00000017319
|
Ppdpf
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr3_-_150064438 | 0.00 |
ENSRNOT00000086933
|
E2f1
|
E2F transcription factor 1 |
| chr15_-_12513931 | 0.00 |
ENSRNOT00000010103
|
Atxn7
|
ataxin 7 |
| chr5_+_140979435 | 0.00 |
ENSRNOT00000056592
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr20_-_1818800 | 0.00 |
ENSRNOT00000000990
|
RT1-M3-1
|
RT1 class Ib, locus M3, gene 1 |
| chr4_-_56114254 | 0.00 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr3_-_176282211 | 0.00 |
ENSRNOT00000013690
|
Bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr3_+_172385672 | 0.00 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr20_+_7484800 | 0.00 |
ENSRNOT00000081604
|
Anks1a
|
ankyrin repeat and sterile alpha motif domain containing 1A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.4 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.2 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0072254 | metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) |
| 0.0 | 0.1 | GO:0070602 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.2 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.1 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:1904339 | cellular response to iron(II) ion(GO:0071282) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0006524 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0034696 | response to prostaglandin F(GO:0034696) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:2000118 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.4 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.0 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.0 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.0 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.0 | 0.1 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNA binding(GO:0030619) U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0004021 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.0 | GO:0008254 | 3'-nucleotidase activity(GO:0008254) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |