Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
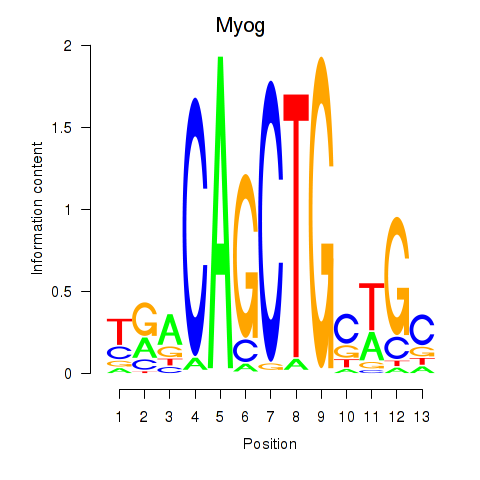
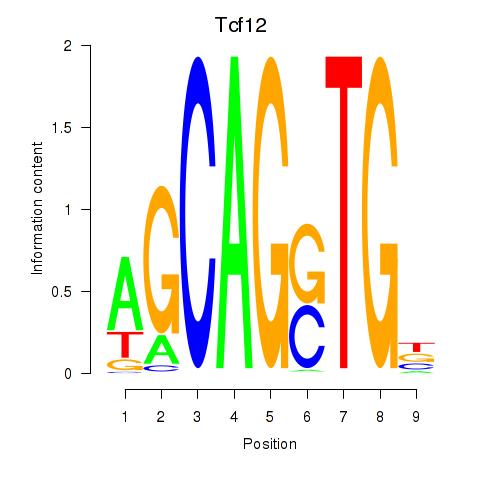
Results for Myog_Tcf12
Z-value: 0.34
Transcription factors associated with Myog_Tcf12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myog
|
ENSRNOG00000030743 | myogenin |
|
Tcf12
|
ENSRNOG00000057754 | transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf12 | rn6_v1_chr8_-_78655856_78655856 | 0.78 | 1.2e-01 | Click! |
Activity profile of Myog_Tcf12 motif
Sorted Z-values of Myog_Tcf12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_115910522 | 0.26 |
ENSRNOT00000076998
ENSRNOT00000067442 |
Arc
|
activity-regulated cytoskeleton-associated protein |
| chr19_-_55510460 | 0.19 |
ENSRNOT00000019820
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr10_+_85301875 | 0.17 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr5_+_63781801 | 0.17 |
ENSRNOT00000008302
|
Nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr11_+_3119885 | 0.16 |
ENSRNOT00000042316
|
Vgll3
|
vestigial-like family member 3 |
| chr7_+_130532435 | 0.16 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr2_-_231521052 | 0.16 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr10_+_71278650 | 0.16 |
ENSRNOT00000092020
|
Synrg
|
synergin, gamma |
| chr2_-_21698937 | 0.15 |
ENSRNOT00000080165
|
AABR07007642.1
|
|
| chr18_+_56193978 | 0.15 |
ENSRNOT00000041533
ENSRNOT00000080177 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr6_-_65319527 | 0.15 |
ENSRNOT00000005618
|
Stxbp6
|
syntaxin binding protein 6 |
| chr3_-_147865393 | 0.14 |
ENSRNOT00000009852
|
Sox12
|
SRY box 12 |
| chr3_+_175144495 | 0.14 |
ENSRNOT00000082601
ENSRNOT00000088026 |
Cdh4
|
cadherin 4 |
| chr17_-_10208360 | 0.12 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr4_-_159697207 | 0.11 |
ENSRNOT00000086440
|
Ccnd2
|
cyclin D2 |
| chr10_+_11393103 | 0.11 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr12_-_25638797 | 0.11 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr9_+_49479023 | 0.11 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chr5_-_58469399 | 0.11 |
ENSRNOT00000013213
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr10_-_64642292 | 0.10 |
ENSRNOT00000084670
|
Abr
|
active BCR-related |
| chr5_+_172364421 | 0.10 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr19_-_58735173 | 0.10 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr15_-_33725188 | 0.10 |
ENSRNOT00000083941
|
Zfhx2
|
zinc finger homeobox 2 |
| chr11_+_71151132 | 0.10 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr12_+_28212333 | 0.10 |
ENSRNOT00000001182
|
Auts2
|
autism susceptibility candidate 2 |
| chr17_+_13670520 | 0.10 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr9_+_53627208 | 0.10 |
ENSRNOT00000083487
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chrX_+_105239840 | 0.09 |
ENSRNOT00000039864
|
Drp2
|
dystrophin related protein 2 |
| chr3_+_11207542 | 0.09 |
ENSRNOT00000013546
|
Prrc2b
|
proline-rich coiled-coil 2B |
| chr4_+_84597323 | 0.09 |
ENSRNOT00000074054
ENSRNOT00000012755 |
Wipf3
|
WAS/WASL interacting protein family, member 3 |
| chr4_-_125929002 | 0.09 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr14_-_112946875 | 0.09 |
ENSRNOT00000081981
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr10_-_110101872 | 0.09 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr10_-_62699723 | 0.08 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr9_+_94425252 | 0.08 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr1_+_192233910 | 0.08 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr10_+_83231238 | 0.08 |
ENSRNOT00000077625
|
Spop
|
speckle type BTB/POZ protein |
| chr7_-_97977135 | 0.08 |
ENSRNOT00000008785
|
Wdyhv1
|
WDYHV motif containing 1 |
| chr9_+_82120059 | 0.08 |
ENSRNOT00000057368
|
Cdk5r2
|
cyclin-dependent kinase 5 regulatory subunit 2 |
| chr8_+_63600663 | 0.08 |
ENSRNOT00000012644
|
Hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr20_+_18780605 | 0.08 |
ENSRNOT00000000754
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr5_+_144634143 | 0.08 |
ENSRNOT00000075558
|
RGD1563072
|
similar to hypothetical protein FLJ38984 |
| chr9_-_82699551 | 0.08 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr13_-_88642011 | 0.08 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr3_-_58181971 | 0.08 |
ENSRNOT00000002076
|
Dlx2
|
distal-less homeobox 2 |
| chr4_-_19413484 | 0.07 |
ENSRNOT00000009489
|
Sema3d
|
semaphorin 3D |
| chr4_+_6282278 | 0.07 |
ENSRNOT00000010349
|
Kmt2c
|
lysine methyltransferase 2C |
| chr2_+_77868412 | 0.07 |
ENSRNOT00000065897
ENSRNOT00000014022 |
Myo10
|
myosin X |
| chr5_-_158439078 | 0.07 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr9_-_80166807 | 0.07 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr16_-_20097287 | 0.07 |
ENSRNOT00000025162
|
Unc13a
|
unc-13 homolog A |
| chr10_-_54467956 | 0.07 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chr15_+_19547871 | 0.07 |
ENSRNOT00000036235
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr7_+_11033317 | 0.07 |
ENSRNOT00000007498
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chr7_-_114590119 | 0.07 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr5_-_151117042 | 0.07 |
ENSRNOT00000066549
|
Fam76a
|
family with sequence similarity 76, member A |
| chr10_+_103713045 | 0.07 |
ENSRNOT00000004351
|
Slc9a3r1
|
SLC9A3 regulator 1 |
| chr18_-_80865584 | 0.07 |
ENSRNOT00000021750
|
Tshz1
|
teashirt zinc finger homeobox 1 |
| chr9_+_111028824 | 0.07 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_210381829 | 0.07 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chr20_+_3558827 | 0.07 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_+_47677720 | 0.07 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr4_+_9966891 | 0.07 |
ENSRNOT00000086609
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr10_-_14299167 | 0.07 |
ENSRNOT00000042066
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr4_-_182844291 | 0.07 |
ENSRNOT00000064320
|
Tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr3_-_7422738 | 0.07 |
ENSRNOT00000088339
|
Gtf3c4
|
general transcription factor IIIC subunit 4 |
| chr17_+_9639330 | 0.07 |
ENSRNOT00000018232
|
Dok3
|
docking protein 3 |
| chr14_+_12218553 | 0.06 |
ENSRNOT00000003237
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr1_-_277768368 | 0.06 |
ENSRNOT00000023113
|
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr7_-_58587787 | 0.06 |
ENSRNOT00000005814
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr1_+_220353356 | 0.06 |
ENSRNOT00000080189
|
Cd248
|
CD248 molecule |
| chr7_+_58366192 | 0.06 |
ENSRNOT00000081891
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr1_-_88920291 | 0.06 |
ENSRNOT00000028306
|
Kirrel2
|
kin of IRRE like 2 (Drosophila) |
| chr17_-_84488480 | 0.06 |
ENSRNOT00000000158
ENSRNOT00000075983 |
Nebl
|
nebulette |
| chr3_-_122947075 | 0.06 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr15_+_110114148 | 0.06 |
ENSRNOT00000006264
|
Itgbl1
|
integrin subunit beta like 1 |
| chr4_+_157538303 | 0.06 |
ENSRNOT00000086418
|
Zfp384
|
zinc finger protein 384 |
| chr18_-_57245666 | 0.06 |
ENSRNOT00000080365
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr1_+_170262156 | 0.06 |
ENSRNOT00000024077
ENSRNOT00000085395 |
Cckbr
|
cholecystokinin B receptor |
| chr17_+_81922329 | 0.06 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr16_-_49820235 | 0.06 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr1_-_190370499 | 0.06 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr1_-_256013495 | 0.06 |
ENSRNOT00000023342
|
Ide
|
insulin degrading enzyme |
| chrX_-_157013443 | 0.06 |
ENSRNOT00000082711
|
Srpk3
|
SRSF protein kinase 3 |
| chr20_-_27083410 | 0.06 |
ENSRNOT00000000432
|
Atoh7
|
atonal bHLH transcription factor 7 |
| chrX_+_53360839 | 0.06 |
ENSRNOT00000091467
ENSRNOT00000034372 ENSRNOT00000081061 |
Dmd
|
dystrophin |
| chr12_-_36398206 | 0.06 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr10_-_92476109 | 0.06 |
ENSRNOT00000089029
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr1_-_64446818 | 0.05 |
ENSRNOT00000081980
|
Myadm
|
myeloid-associated differentiation marker |
| chr17_+_9596957 | 0.05 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr13_-_51784639 | 0.05 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr7_-_2712723 | 0.05 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr18_-_14016713 | 0.05 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr13_-_76049363 | 0.05 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr1_+_220362064 | 0.05 |
ENSRNOT00000074361
|
LOC108348067
|
endosialin |
| chr1_+_101214593 | 0.05 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr1_+_79989019 | 0.05 |
ENSRNOT00000020428
|
Dmpk
|
dystrophia myotonica-protein kinase |
| chr17_-_80320681 | 0.05 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr15_+_120372 | 0.05 |
ENSRNOT00000007828
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr10_-_4910305 | 0.05 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr10_-_31359699 | 0.05 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr18_-_38088457 | 0.05 |
ENSRNOT00000077814
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr5_+_48274477 | 0.05 |
ENSRNOT00000075992
ENSRNOT00000041271 |
Ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr1_+_80000165 | 0.05 |
ENSRNOT00000084912
|
Six5
|
SIX homeobox 5 |
| chr3_+_41019898 | 0.05 |
ENSRNOT00000007335
|
Kcnj3
|
potassium voltage-gated channel subfamily J member 3 |
| chrX_+_88298266 | 0.05 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr10_-_62814520 | 0.05 |
ENSRNOT00000019410
|
Ssh2
|
slingshot protein phosphatase 2 |
| chr16_+_20740826 | 0.05 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr8_+_68526093 | 0.05 |
ENSRNOT00000011385
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chrX_+_80213332 | 0.05 |
ENSRNOT00000042827
|
Sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr5_-_19559244 | 0.05 |
ENSRNOT00000014289
ENSRNOT00000089666 |
Nsmaf
|
neutral sphingomyelinase activation associated factor |
| chr4_-_153593773 | 0.05 |
ENSRNOT00000082982
|
Mical3
|
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
| chr3_-_7796385 | 0.05 |
ENSRNOT00000082489
|
Ntng2
|
netrin G2 |
| chr13_+_52662996 | 0.05 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr7_-_121029754 | 0.05 |
ENSRNOT00000004703
|
Nptxr
|
neuronal pentraxin receptor |
| chr1_-_215846911 | 0.05 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr13_+_34400170 | 0.05 |
ENSRNOT00000061516
ENSRNOT00000061515 ENSRNOT00000061513 ENSRNOT00000084506 ENSRNOT00000086641 |
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr3_+_177164359 | 0.05 |
ENSRNOT00000066837
|
RGD1561282
|
RGD1561282 |
| chr5_+_58505500 | 0.05 |
ENSRNOT00000043216
|
Unc13b
|
unc-13 homolog B |
| chr2_-_31753528 | 0.05 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr15_-_37383277 | 0.05 |
ENSRNOT00000011711
|
Gjb2
|
gap junction protein, beta 2 |
| chr11_+_45888221 | 0.05 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr10_-_55965216 | 0.05 |
ENSRNOT00000057058
|
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr2_+_11658568 | 0.05 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr7_-_140580783 | 0.05 |
ENSRNOT00000087327
|
Dhh
|
desert hedgehog |
| chr1_+_278557792 | 0.05 |
ENSRNOT00000023414
|
Atrnl1
|
attractin like 1 |
| chr10_+_89339029 | 0.05 |
ENSRNOT00000031208
|
Rundc1
|
RUN domain containing 1 |
| chr3_+_11392791 | 0.04 |
ENSRNOT00000064792
|
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr16_+_32457521 | 0.04 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chrX_-_159841072 | 0.04 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr1_+_248228496 | 0.04 |
ENSRNOT00000015406
|
Uhrf2
|
ubiquitin like with PHD and ring finger domains 2 |
| chr14_-_112946204 | 0.04 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr5_-_153625869 | 0.04 |
ENSRNOT00000024464
|
Clic4
|
chloride intracellular channel 4 |
| chr1_-_281874456 | 0.04 |
ENSRNOT00000084760
ENSRNOT00000050617 |
Cacul1
|
CDK2-associated, cullin domain 1 |
| chr19_-_53688597 | 0.04 |
ENSRNOT00000074448
|
Zcchc14
|
zinc finger CCHC-type containing 14 |
| chr4_-_117268178 | 0.04 |
ENSRNOT00000043201
ENSRNOT00000084049 |
Fbxo41
|
F-box protein 41 |
| chr1_-_141188031 | 0.04 |
ENSRNOT00000044567
|
Polg
|
DNA polymerase gamma, catalytic subunit |
| chr10_+_47019326 | 0.04 |
ENSRNOT00000006984
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr6_-_138508753 | 0.04 |
ENSRNOT00000006888
|
Ighm
|
immunoglobulin heavy constant mu |
| chrX_+_70596901 | 0.04 |
ENSRNOT00000088114
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr2_+_149494330 | 0.04 |
ENSRNOT00000074764
ENSRNOT00000089865 |
Med12l
|
mediator complex subunit 12-like |
| chr1_+_255185629 | 0.04 |
ENSRNOT00000083002
|
Hectd2
|
HECT domain E3 ubiquitin protein ligase 2 |
| chrX_+_82143789 | 0.04 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr11_-_88273254 | 0.04 |
ENSRNOT00000002533
|
Mapk1
|
mitogen activated protein kinase 1 |
| chr2_+_210977938 | 0.04 |
ENSRNOT00000074725
|
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr16_+_46731403 | 0.04 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr15_+_23792931 | 0.04 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr3_-_107760550 | 0.04 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr4_+_120672152 | 0.04 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr12_+_7081895 | 0.04 |
ENSRNOT00000047163
|
Hmgb1
|
high mobility group box 1 |
| chrX_-_123515720 | 0.04 |
ENSRNOT00000092343
|
Nkrf
|
NFKB repressing factor |
| chr2_-_140618405 | 0.04 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr12_+_6403940 | 0.04 |
ENSRNOT00000083484
|
B3glct
|
beta 3-glucosyltransferase |
| chr19_+_54060622 | 0.04 |
ENSRNOT00000023607
|
Gse1
|
Gse1 coiled-coil protein |
| chr2_+_198772937 | 0.04 |
ENSRNOT00000028812
|
Itga10
|
integrin subunit alpha 10 |
| chr10_-_67751002 | 0.04 |
ENSRNOT00000000254
|
RGD1310429
|
similar to Protein Njmu-R1 |
| chr3_-_110556808 | 0.04 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr3_+_81134505 | 0.04 |
ENSRNOT00000067664
|
Phf21a
|
PHD finger protein 21A |
| chr5_-_16140896 | 0.04 |
ENSRNOT00000029503
|
Xkr4
|
XK related 4 |
| chr10_+_74959285 | 0.04 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr13_+_89718066 | 0.04 |
ENSRNOT00000005147
|
Dedd
|
death effector domain-containing |
| chr8_-_44327551 | 0.04 |
ENSRNOT00000083939
|
Gramd1b
|
GRAM domain containing 1B |
| chr5_-_79874671 | 0.04 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr15_-_33752665 | 0.04 |
ENSRNOT00000034102
|
Zfhx2
|
zinc finger homeobox 2 |
| chr8_-_62616828 | 0.04 |
ENSRNOT00000068340
|
Arid3b
|
AT-rich interaction domain 3B |
| chr7_+_11383116 | 0.04 |
ENSRNOT00000066348
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr13_+_44475970 | 0.04 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chr16_+_10727571 | 0.04 |
ENSRNOT00000084422
|
Mmrn2
|
multimerin 2 |
| chr4_-_83137527 | 0.04 |
ENSRNOT00000039580
|
Jazf1
|
JAZF zinc finger 1 |
| chr15_-_90175802 | 0.04 |
ENSRNOT00000013342
|
Spry2
|
sprouty RTK signaling antagonist 2 |
| chr1_+_78800754 | 0.04 |
ENSRNOT00000084601
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr2_+_196334626 | 0.04 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr3_+_79918969 | 0.04 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr1_-_219544328 | 0.04 |
ENSRNOT00000025847
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr5_+_1417478 | 0.04 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chr20_-_5865775 | 0.04 |
ENSRNOT00000000612
ENSRNOT00000092641 |
Srpk1
|
SRSF protein kinase 1 |
| chr1_-_277939039 | 0.04 |
ENSRNOT00000079579
ENSRNOT00000086726 |
Ablim1
|
actin-binding LIM protein 1 |
| chr5_+_151362019 | 0.04 |
ENSRNOT00000077083
ENSRNOT00000067801 |
Wasf2
|
WAS protein family, member 2 |
| chr18_+_32594958 | 0.04 |
ENSRNOT00000018511
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr10_-_31419235 | 0.03 |
ENSRNOT00000059496
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr8_+_39960542 | 0.03 |
ENSRNOT00000013370
|
Msantd2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr10_-_85435016 | 0.03 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr19_+_784618 | 0.03 |
ENSRNOT00000014749
|
Cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr12_+_47590154 | 0.03 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr3_-_156340913 | 0.03 |
ENSRNOT00000021452
|
Mafb
|
MAF bZIP transcription factor B |
| chr2_-_186245163 | 0.03 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr8_-_96132634 | 0.03 |
ENSRNOT00000041655
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr10_-_67285617 | 0.03 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chrX_+_1297099 | 0.03 |
ENSRNOT00000013522
|
Elk1
|
ELK1, ETS transcription factor |
| chr8_+_27777179 | 0.03 |
ENSRNOT00000009825
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 |
| chr13_-_73558912 | 0.03 |
ENSRNOT00000093655
|
Cep350
|
centrosomal protein 350 |
| chr1_+_154377447 | 0.03 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr20_-_2701637 | 0.03 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr17_+_44758460 | 0.03 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chr3_-_72289310 | 0.03 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr7_-_98098268 | 0.03 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chrX_+_68774699 | 0.03 |
ENSRNOT00000081662
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr16_+_19686630 | 0.03 |
ENSRNOT00000081321
|
Myo9b
|
myosin IXb |
| chr15_+_61069581 | 0.03 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr11_-_90406797 | 0.03 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myog_Tcf12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:1900625 | regulation of mast cell cytokine production(GO:0032763) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) NMDA selective glutamate receptor signaling pathway(GO:0098989) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0072218 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:1904722 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0021586 | pons maturation(GO:0021586) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:2000328 | regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.0 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.0 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.0 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.0 | GO:0070563 | cell migration involved in endocardial cushion formation(GO:0003273) negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.0 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.0 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |