Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
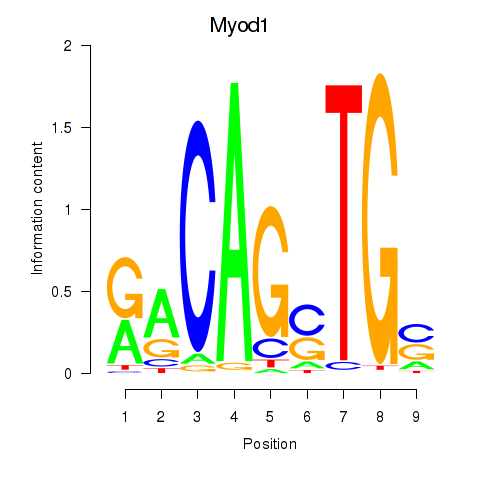
Results for Myod1
Z-value: 0.37
Transcription factors associated with Myod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myod1
|
ENSRNOG00000011306 | myogenic differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myod1 | rn6_v1_chr1_+_102396538_102396538 | 0.17 | 7.8e-01 | Click! |
Activity profile of Myod1 motif
Sorted Z-values of Myod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_94425252 | 0.14 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr10_+_70262361 | 0.10 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chr7_+_130532435 | 0.10 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr2_-_231521052 | 0.10 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr1_-_88920291 | 0.09 |
ENSRNOT00000028306
|
Kirrel2
|
kin of IRRE like 2 (Drosophila) |
| chr1_+_241594565 | 0.09 |
ENSRNOT00000020123
|
Apba1
|
amyloid beta precursor protein binding family A member 1 |
| chr9_+_111028824 | 0.09 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_110556808 | 0.08 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr5_-_58469399 | 0.08 |
ENSRNOT00000013213
|
Pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr11_-_88038518 | 0.08 |
ENSRNOT00000051991
|
Rimbp3
|
RIMS binding protein 3 |
| chr8_-_120446455 | 0.08 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr9_+_16749809 | 0.08 |
ENSRNOT00000028786
|
Cul9
|
cullin 9 |
| chr10_+_71278650 | 0.07 |
ENSRNOT00000092020
|
Synrg
|
synergin, gamma |
| chr6_+_27328406 | 0.07 |
ENSRNOT00000091159
ENSRNOT00000044278 |
Otof
|
otoferlin |
| chr1_+_80321585 | 0.07 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr12_+_31530699 | 0.07 |
ENSRNOT00000033379
ENSRNOT00000067867 |
Rimbp2
|
RIMS binding protein 2 |
| chr16_+_46731403 | 0.07 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr12_+_660011 | 0.07 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr7_+_11383116 | 0.07 |
ENSRNOT00000066348
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr20_+_3558827 | 0.07 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr10_+_11393103 | 0.07 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chrX_+_88298266 | 0.07 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr19_-_55510460 | 0.07 |
ENSRNOT00000019820
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr1_-_75154325 | 0.07 |
ENSRNOT00000074520
|
RGD1564801
|
similar to hepatic multiple inositol polyphosphate phosphatase |
| chr1_+_192233910 | 0.07 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr10_+_90731865 | 0.06 |
ENSRNOT00000064429
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr4_-_147893992 | 0.06 |
ENSRNOT00000032158
|
Plxnd1
|
plexin D1 |
| chr2_-_219262901 | 0.06 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr10_+_85301875 | 0.06 |
ENSRNOT00000080935
|
Socs7
|
suppressor of cytokine signaling 7 |
| chr10_-_55965216 | 0.06 |
ENSRNOT00000057058
|
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr10_-_89338739 | 0.06 |
ENSRNOT00000073923
|
Ptges3l
|
prostaglandin E synthase 3 like |
| chr15_-_33725188 | 0.06 |
ENSRNOT00000083941
|
Zfhx2
|
zinc finger homeobox 2 |
| chrX_-_26376467 | 0.06 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr18_+_44468784 | 0.05 |
ENSRNOT00000031812
|
Dmxl1
|
Dmx-like 1 |
| chr14_+_104250617 | 0.05 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr2_+_11658568 | 0.05 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr17_-_89163113 | 0.05 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr4_-_19413484 | 0.05 |
ENSRNOT00000009489
|
Sema3d
|
semaphorin 3D |
| chr7_-_50638798 | 0.05 |
ENSRNOT00000048880
|
Syt1
|
synaptotagmin 1 |
| chr11_-_68349238 | 0.05 |
ENSRNOT00000030074
|
Sema5b
|
semaphorin 5B |
| chr11_-_83926524 | 0.05 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr5_+_151362019 | 0.05 |
ENSRNOT00000077083
ENSRNOT00000067801 |
Wasf2
|
WAS protein family, member 2 |
| chr3_-_94418711 | 0.05 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr18_+_14756684 | 0.05 |
ENSRNOT00000076085
ENSRNOT00000076129 |
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_+_251863069 | 0.05 |
ENSRNOT00000036282
|
Syde2
|
synapse defective Rho GTPase homolog 2 |
| chr20_-_8202924 | 0.05 |
ENSRNOT00000071399
|
Tmem217
|
transmembrane protein 217 |
| chr13_-_76049363 | 0.04 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr4_-_88649216 | 0.04 |
ENSRNOT00000058626
|
Herc6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr5_-_16140896 | 0.04 |
ENSRNOT00000029503
|
Xkr4
|
XK related 4 |
| chr1_+_167374182 | 0.04 |
ENSRNOT00000092882
|
Stim1
|
stromal interaction molecule 1 |
| chr3_-_66885085 | 0.04 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr10_-_62699723 | 0.04 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr11_-_45510961 | 0.04 |
ENSRNOT00000002238
|
Tomm70
|
translocase of outer mitochondrial membrane 70 |
| chr1_-_80331626 | 0.04 |
ENSRNOT00000022577
|
AABR07002677.1
|
|
| chr1_+_199287710 | 0.04 |
ENSRNOT00000082121
|
Stx4
|
syntaxin 4 |
| chr1_+_104635989 | 0.04 |
ENSRNOT00000078477
|
Nav2
|
neuron navigator 2 |
| chr2_+_189423559 | 0.04 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr2_+_187988925 | 0.04 |
ENSRNOT00000093033
ENSRNOT00000093016 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr11_+_45888221 | 0.04 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr17_+_75352389 | 0.04 |
ENSRNOT00000023436
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr8_+_68526093 | 0.04 |
ENSRNOT00000011385
|
Aagab
|
alpha- and gamma-adaptin binding protein |
| chr1_-_220265772 | 0.04 |
ENSRNOT00000027119
|
Npas4
|
neuronal PAS domain protein 4 |
| chr11_-_11214141 | 0.04 |
ENSRNOT00000065748
|
Robo2
|
roundabout guidance receptor 2 |
| chr1_-_54748763 | 0.04 |
ENSRNOT00000074549
|
LOC100911027
|
protein MAL2-like |
| chr13_+_44475970 | 0.04 |
ENSRNOT00000024602
ENSRNOT00000091645 |
Ccnt2
|
cyclin T2 |
| chr20_-_47306318 | 0.04 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr2_-_186245163 | 0.04 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr14_-_81023682 | 0.04 |
ENSRNOT00000081570
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr5_+_144634143 | 0.04 |
ENSRNOT00000075558
|
RGD1563072
|
similar to hypothetical protein FLJ38984 |
| chr15_+_4240203 | 0.04 |
ENSRNOT00000010178
|
Mss51
|
MSS51 mitochondrial translational activator |
| chr10_+_83231238 | 0.04 |
ENSRNOT00000077625
|
Spop
|
speckle type BTB/POZ protein |
| chr7_+_141702038 | 0.04 |
ENSRNOT00000086577
|
Dip2b
|
disco-interacting protein 2 homolog B |
| chr1_+_266952561 | 0.03 |
ENSRNOT00000076452
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr16_+_19686630 | 0.03 |
ENSRNOT00000081321
|
Myo9b
|
myosin IXb |
| chr12_-_44279002 | 0.03 |
ENSRNOT00000064900
|
Fbxo21
|
F-box protein 21 |
| chr2_+_179952227 | 0.03 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr15_+_120372 | 0.03 |
ENSRNOT00000007828
|
Dlg5
|
discs large MAGUK scaffold protein 5 |
| chr1_+_260093641 | 0.03 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr5_+_3955227 | 0.03 |
ENSRNOT00000009908
|
Msc
|
musculin |
| chr10_-_15928169 | 0.03 |
ENSRNOT00000028069
|
Nsg2
|
neuron specific gene family member 2 |
| chr8_-_87419564 | 0.03 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr9_-_82382272 | 0.03 |
ENSRNOT00000025627
|
Abcb6
|
ATP-binding cassette, subfamily B (MDR/TAP), member 6 |
| chr18_-_14016713 | 0.03 |
ENSRNOT00000041125
|
Nol4
|
nucleolar protein 4 |
| chr2_+_77868412 | 0.03 |
ENSRNOT00000065897
ENSRNOT00000014022 |
Myo10
|
myosin X |
| chr3_+_5624506 | 0.03 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr10_+_92628356 | 0.03 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr2_-_94730978 | 0.03 |
ENSRNOT00000087598
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr4_-_129619142 | 0.03 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr13_-_111581018 | 0.03 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chrX_+_71335491 | 0.03 |
ENSRNOT00000076003
|
Nono
|
non-POU domain containing, octamer-binding |
| chr1_-_88690534 | 0.03 |
ENSRNOT00000072570
|
Ovol3
|
ovo-like zinc finger 3 |
| chr3_-_81304181 | 0.03 |
ENSRNOT00000079746
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chrX_-_136150209 | 0.03 |
ENSRNOT00000048498
|
Enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr4_+_6282278 | 0.03 |
ENSRNOT00000010349
|
Kmt2c
|
lysine methyltransferase 2C |
| chr10_+_44271587 | 0.03 |
ENSRNOT00000047700
|
Trim58
|
tripartite motif-containing 58 |
| chr8_-_96132634 | 0.03 |
ENSRNOT00000041655
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chrX_+_107695491 | 0.03 |
ENSRNOT00000071973
|
Fam199x
|
family with sequence similarity 199, X-linked |
| chr6_-_50846965 | 0.03 |
ENSRNOT00000087300
|
Slc26a4
|
solute carrier family 26 member 4 |
| chr2_-_181026024 | 0.03 |
ENSRNOT00000064930
|
Gucy1b3
|
guanylate cyclase 1 soluble subunit beta 3 |
| chr10_-_29450644 | 0.03 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr10_-_95934345 | 0.03 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr7_-_36000906 | 0.03 |
ENSRNOT00000011178
|
Plxnc1
|
plexin C1 |
| chr3_+_172155496 | 0.02 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
| chr8_-_62616828 | 0.02 |
ENSRNOT00000068340
|
Arid3b
|
AT-rich interaction domain 3B |
| chr10_+_34185898 | 0.02 |
ENSRNOT00000003339
|
Trim7
|
tripartite motif-containing 7 |
| chr9_-_71445739 | 0.02 |
ENSRNOT00000019698
|
Fzd5
|
frizzled class receptor 5 |
| chr9_-_32019205 | 0.02 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr1_+_128924966 | 0.02 |
ENSRNOT00000019267
|
Igf1r
|
insulin-like growth factor 1 receptor |
| chr18_+_60392376 | 0.02 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr12_-_36398206 | 0.02 |
ENSRNOT00000090944
ENSRNOT00000058492 |
Tmem132b
|
transmembrane protein 132B |
| chr6_-_99783047 | 0.02 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chr3_-_122947075 | 0.02 |
ENSRNOT00000082369
|
Pced1a
|
PC-esterase domain containing 1A |
| chr1_-_128695995 | 0.02 |
ENSRNOT00000077020
|
Synm
|
synemin |
| chr2_+_196594303 | 0.02 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr6_-_72462657 | 0.02 |
ENSRNOT00000088720
ENSRNOT00000077578 |
Strn3
|
striatin 3 |
| chr15_-_8931983 | 0.02 |
ENSRNOT00000085322
|
Thrb
|
thyroid hormone receptor beta |
| chr6_+_127207732 | 0.02 |
ENSRNOT00000071976
|
RGD1309028
|
similar to RIKEN cDNA A830059I20 |
| chr9_+_111028575 | 0.02 |
ENSRNOT00000043451
|
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_+_50599060 | 0.02 |
ENSRNOT00000058420
|
AABR07038477.1
|
|
| chr1_+_215609036 | 0.02 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr3_-_123723524 | 0.02 |
ENSRNOT00000080351
|
Cenpb
|
centromere protein B |
| chr14_-_82287706 | 0.02 |
ENSRNOT00000080695
|
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr19_+_54060622 | 0.02 |
ENSRNOT00000023607
|
Gse1
|
Gse1 coiled-coil protein |
| chr4_-_62438958 | 0.02 |
ENSRNOT00000014010
|
Wdr91
|
WD repeat domain 91 |
| chr1_+_15642153 | 0.02 |
ENSRNOT00000079845
|
Map7
|
microtubule-associated protein 7 |
| chr7_-_98098268 | 0.02 |
ENSRNOT00000010361
|
Fbxo32
|
F-box protein 32 |
| chr8_+_65733400 | 0.02 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr19_+_17290178 | 0.02 |
ENSRNOT00000060865
|
Aktip
|
AKT interacting protein |
| chr5_+_58937615 | 0.02 |
ENSRNOT00000080909
|
Tesk1
|
testis-specific kinase 1 |
| chr7_+_63922879 | 0.02 |
ENSRNOT00000043581
|
RGD1565498
|
similar to Hypothetical protein LOC270802 |
| chr7_+_71065197 | 0.02 |
ENSRNOT00000051075
|
Rdh16
|
retinol dehydrogenase 16 (all-trans) |
| chr4_-_178168690 | 0.02 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr6_-_128727374 | 0.02 |
ENSRNOT00000082152
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr4_+_65637407 | 0.01 |
ENSRNOT00000047954
|
Trim24
|
tripartite motif-containing 24 |
| chr1_-_213650247 | 0.01 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr1_+_265298868 | 0.01 |
ENSRNOT00000023278
|
Dpcd
|
deleted in primary ciliary dyskinesia |
| chr4_+_6559545 | 0.01 |
ENSRNOT00000067183
|
Prkag2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr13_-_110642754 | 0.01 |
ENSRNOT00000029039
|
LOC100362350
|
hydroxysteroid 17-beta dehydrogenase 6-like |
| chr1_-_31055453 | 0.01 |
ENSRNOT00000031083
|
Soga3
|
SOGA family member 3 |
| chr10_+_91254058 | 0.01 |
ENSRNOT00000087218
ENSRNOT00000065373 |
Fmnl1
|
formin-like 1 |
| chr9_+_88357556 | 0.01 |
ENSRNOT00000020669
|
Col4a3
|
collagen type IV alpha 3 chain |
| chr15_+_33555640 | 0.01 |
ENSRNOT00000021096
|
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr17_+_31441630 | 0.01 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr19_+_52032886 | 0.01 |
ENSRNOT00000019923
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr1_-_215846911 | 0.01 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr5_-_101006375 | 0.01 |
ENSRNOT00000014008
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr3_+_13838304 | 0.01 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr5_-_113578928 | 0.01 |
ENSRNOT00000010621
|
Plaa
|
phospholipase A2, activating protein |
| chr19_+_57484634 | 0.01 |
ENSRNOT00000025695
|
Arv1
|
ARV1 homolog, fatty acid homeostasis modulator |
| chr1_+_215609645 | 0.01 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr13_-_73704480 | 0.01 |
ENSRNOT00000005296
|
Tor1aip1
|
torsin 1A interacting protein 1 |
| chr8_-_122987191 | 0.01 |
ENSRNOT00000033976
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr16_-_40025401 | 0.01 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr10_-_8654892 | 0.01 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr6_-_105518725 | 0.01 |
ENSRNOT00000009693
ENSRNOT00000083488 |
Map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr15_+_2526368 | 0.01 |
ENSRNOT00000048713
ENSRNOT00000074803 |
Dusp13
Dusp13
|
dual specificity phosphatase 13 dual specificity phosphatase 13 |
| chrX_-_112328642 | 0.01 |
ENSRNOT00000083150
|
Psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr19_-_55434252 | 0.01 |
ENSRNOT00000045052
|
Pabpn1l
|
poly(A)binding protein nuclear 1-like |
| chr11_-_82860977 | 0.01 |
ENSRNOT00000002382
|
Polr2h
|
RNA polymerase II subunit H |
| chr2_-_43999107 | 0.01 |
ENSRNOT00000065473
|
LOC100912399
|
mitogen-activated protein kinase kinase kinase 1-like |
| chr13_+_51958834 | 0.01 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr8_-_116469915 | 0.01 |
ENSRNOT00000024193
|
Sema3f
|
semaphorin 3F |
| chr7_+_94130852 | 0.01 |
ENSRNOT00000011485
|
Mal2
|
mal, T-cell differentiation protein 2 |
| chr10_-_109827143 | 0.01 |
ENSRNOT00000054948
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr8_-_33017854 | 0.01 |
ENSRNOT00000011386
|
Barx2
|
BARX homeobox 2 |
| chr2_+_197720259 | 0.01 |
ENSRNOT00000070919
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr5_+_48313599 | 0.01 |
ENSRNOT00000081825
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr10_-_56511583 | 0.01 |
ENSRNOT00000021402
|
LOC497940
|
similar to RIKEN cDNA 2810408A11 |
| chr2_-_184289126 | 0.01 |
ENSRNOT00000081678
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr1_-_44474674 | 0.01 |
ENSRNOT00000078768
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr5_+_61657507 | 0.01 |
ENSRNOT00000013228
|
Tmod1
|
tropomodulin 1 |
| chr1_+_101214593 | 0.01 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr1_+_220353356 | 0.01 |
ENSRNOT00000080189
|
Cd248
|
CD248 molecule |
| chr13_+_52889737 | 0.01 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr13_+_60435946 | 0.01 |
ENSRNOT00000004342
|
B3galt2
|
Beta-1,3-galactosyltransferase 2 |
| chr1_-_72464492 | 0.01 |
ENSRNOT00000068550
|
Nat14
|
N-acetyltransferase 14 |
| chr16_+_32457521 | 0.01 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr4_+_113970079 | 0.01 |
ENSRNOT00000079547
|
Rtkn
|
rhotekin |
| chrX_-_111191932 | 0.01 |
ENSRNOT00000088050
ENSRNOT00000083613 |
Morc4
|
MORC family CW-type zinc finger 4 |
| chr2_+_3400977 | 0.01 |
ENSRNOT00000093593
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr5_+_28850950 | 0.01 |
ENSRNOT00000009759
|
Tmem64
|
transmembrane protein 64 |
| chr5_-_59025631 | 0.01 |
ENSRNOT00000049000
ENSRNOT00000022801 |
Tpm2
|
tropomyosin 2, beta |
| chr17_+_9639330 | 0.01 |
ENSRNOT00000018232
|
Dok3
|
docking protein 3 |
| chr13_+_52662996 | 0.01 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr8_+_130416355 | 0.01 |
ENSRNOT00000026234
|
Klhl40
|
kelch-like family member 40 |
| chr9_-_82419288 | 0.00 |
ENSRNOT00000004797
|
Tuba4a
|
tubulin, alpha 4A |
| chr7_+_79138925 | 0.00 |
ENSRNOT00000047511
|
AABR07057586.1
|
|
| chr15_-_108120279 | 0.00 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr9_-_82336806 | 0.00 |
ENSRNOT00000024667
|
Slc23a3
|
solute carrier family 23, member 3 |
| chr7_+_102586313 | 0.00 |
ENSRNOT00000006188
|
Myc
|
myelocytomatosis oncogene |
| chr5_-_79317110 | 0.00 |
ENSRNOT00000082513
ENSRNOT00000002309 |
Whrn
|
whirlin |
| chr2_+_189922996 | 0.00 |
ENSRNOT00000016499
|
S100a16
|
S100 calcium binding protein A16 |
| chr5_+_18901039 | 0.00 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chrX_+_86126157 | 0.00 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr13_+_99173484 | 0.00 |
ENSRNOT00000080574
ENSRNOT00000088654 |
Lefty2
|
Left-right determination factor 2 |
| chr5_+_147476221 | 0.00 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr2_+_58462949 | 0.00 |
ENSRNOT00000080618
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr2_-_118745766 | 0.00 |
ENSRNOT00000013858
|
Zmat3
|
zinc finger, matrin type 3 |
| chr7_-_144837583 | 0.00 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr7_+_141054494 | 0.00 |
ENSRNOT00000088176
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.0 | GO:1902568 | regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |