Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
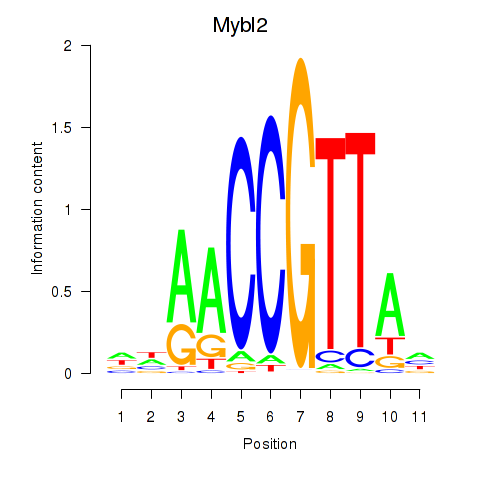
Results for Mybl2
Z-value: 0.50
Transcription factors associated with Mybl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl2
|
ENSRNOG00000007805 | myeloblastosis oncogene-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl2 | rn6_v1_chr3_+_159421671_159421671 | 0.24 | 7.0e-01 | Click! |
Activity profile of Mybl2 motif
Sorted Z-values of Mybl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_88964525 | 0.35 |
ENSRNOT00000068236
|
Daw1
|
dynein assembly factor with WD repeats 1 |
| chr13_+_83681322 | 0.23 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr2_+_58724855 | 0.22 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr8_+_52829085 | 0.22 |
ENSRNOT00000007754
|
RGD1563941
|
similar to hypothetical protein FLJ20010 |
| chr1_-_166943592 | 0.21 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr10_+_14828597 | 0.17 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chr10_-_56531483 | 0.15 |
ENSRNOT00000022343
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr2_+_197682000 | 0.15 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr2_+_127538659 | 0.15 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr9_-_30251388 | 0.15 |
ENSRNOT00000035033
|
Sdhaf4
|
succinate dehydrogenase complex assembly factor 4 |
| chr3_+_147894558 | 0.15 |
ENSRNOT00000055415
|
LOC502684
|
hypothetical protein LOC502684 |
| chr13_-_102643223 | 0.14 |
ENSRNOT00000003155
|
Hlx
|
H2.0-like homeobox |
| chr9_-_94634109 | 0.14 |
ENSRNOT00000081131
|
AABR07068274.1
|
|
| chrX_-_136807885 | 0.14 |
ENSRNOT00000010325
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr20_+_3677474 | 0.14 |
ENSRNOT00000047663
|
Mt1m
|
metallothionein 1M |
| chr18_-_40452456 | 0.13 |
ENSRNOT00000004747
|
Mospd4
|
motile sperm domain containing 4 |
| chr1_+_142087208 | 0.13 |
ENSRNOT00000017532
|
Prc1
|
protein regulator of cytokinesis 1 |
| chr20_+_46044892 | 0.13 |
ENSRNOT00000057187
|
Ak9
|
adenylate kinase 9 |
| chr20_+_14577166 | 0.13 |
ENSRNOT00000085583
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr7_+_2479311 | 0.12 |
ENSRNOT00000003787
|
Ptges3
|
prostaglandin E synthase 3 |
| chr9_+_98313632 | 0.12 |
ENSRNOT00000027012
|
Ramp1
|
receptor activity modifying protein 1 |
| chr3_-_14538241 | 0.12 |
ENSRNOT00000025904
|
Stom
|
stomatin |
| chr5_-_127273656 | 0.12 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr3_-_79728879 | 0.11 |
ENSRNOT00000012425
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr14_+_85351358 | 0.11 |
ENSRNOT00000031914
|
Rhbdd3
|
rhomboid domain containing 3 |
| chr1_+_72580424 | 0.11 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr7_+_76980040 | 0.11 |
ENSRNOT00000050726
|
LOC108350501
|
40S ribosomal protein S29 |
| chr19_+_37795658 | 0.10 |
ENSRNOT00000025686
|
Tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr17_-_13393243 | 0.10 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr15_-_61873406 | 0.10 |
ENSRNOT00000045115
|
LOC100360244
|
LRRGT00053-like |
| chr16_+_72401887 | 0.10 |
ENSRNOT00000074449
|
LOC100910163
|
uncharacterized LOC100910163 |
| chr6_+_99817431 | 0.10 |
ENSRNOT00000009621
|
Churc1
|
churchill domain containing 1 |
| chr1_-_162713610 | 0.09 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr10_+_12929471 | 0.09 |
ENSRNOT00000036563
|
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr1_+_226234074 | 0.09 |
ENSRNOT00000027834
|
Fads1
|
fatty acid desaturase 1 |
| chr9_-_82477136 | 0.09 |
ENSRNOT00000026753
ENSRNOT00000089555 |
Resp18
|
regulated endocrine-specific protein 18 |
| chr1_+_94718402 | 0.09 |
ENSRNOT00000046035
|
Uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr6_+_135139292 | 0.09 |
ENSRNOT00000043508
|
Rpl35al1
|
ribosomal protein L35a-like 1 |
| chr9_+_95256627 | 0.09 |
ENSRNOT00000025291
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr4_-_179904844 | 0.09 |
ENSRNOT00000085386
|
Lmntd1
|
lamin tail domain containing 1 |
| chr17_-_2705123 | 0.08 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr3_+_119776925 | 0.08 |
ENSRNOT00000018549
|
Dusp2
|
dual specificity phosphatase 2 |
| chr13_-_89565813 | 0.08 |
ENSRNOT00000004347
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr14_-_81339526 | 0.08 |
ENSRNOT00000015894
|
Grk4
|
G protein-coupled receptor kinase 4 |
| chr20_-_28814636 | 0.08 |
ENSRNOT00000086030
|
Sept10
|
septin 10 |
| chr3_-_151688149 | 0.08 |
ENSRNOT00000072945
|
Nfs1
|
NFS1 cysteine desulfurase |
| chr8_-_116297850 | 0.08 |
ENSRNOT00000031739
ENSRNOT00000082320 |
Cyb561d2
|
cytochrome b561 family, member D2 |
| chr19_+_16415636 | 0.08 |
ENSRNOT00000089975
|
Irx5
|
iroquois homeobox 5 |
| chr4_-_38240848 | 0.08 |
ENSRNOT00000007567
|
Ndufa4
|
NADH:ubiquinone oxidoreductase subunit A4 |
| chr6_-_135049728 | 0.08 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
| chr11_+_36736586 | 0.08 |
ENSRNOT00000082098
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr8_-_97083120 | 0.08 |
ENSRNOT00000018603
|
Tmed3
|
transmembrane p24 trafficking protein 3 |
| chr10_-_13168217 | 0.08 |
ENSRNOT00000087768
|
Elob
|
elongin B |
| chr20_+_3422461 | 0.08 |
ENSRNOT00000084917
ENSRNOT00000079854 |
Tubb5
|
tubulin, beta 5 class I |
| chr10_-_58924137 | 0.08 |
ENSRNOT00000066740
|
Tekt1
|
tektin 1 |
| chr19_+_25946979 | 0.08 |
ENSRNOT00000004027
|
Gadd45gip1
|
GADD45G interacting protein 1 |
| chr5_-_152070984 | 0.08 |
ENSRNOT00000081107
|
RGD1562725
|
similar to 40S ribosomal protein S20 |
| chr3_-_81282157 | 0.07 |
ENSRNOT00000051258
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr14_-_18704059 | 0.07 |
ENSRNOT00000081455
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr3_+_23301455 | 0.07 |
ENSRNOT00000019405
|
Arpc5l
|
actin related protein 2/3 complex, subunit 5-like |
| chr4_+_92431710 | 0.07 |
ENSRNOT00000049438
|
LOC108350839
|
high mobility group protein B1-like |
| chr10_-_59112788 | 0.07 |
ENSRNOT00000041886
|
Spns3
|
SPNS sphingolipid transporter 3 |
| chr8_+_128824508 | 0.07 |
ENSRNOT00000025343
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr3_+_152222726 | 0.07 |
ENSRNOT00000072464
|
LOC100910944
|
reactive oxygen species modulator 1-like |
| chr12_+_47024442 | 0.07 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr14_-_84937725 | 0.07 |
ENSRNOT00000083839
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr8_-_56393233 | 0.07 |
ENSRNOT00000016263
|
Fdx1
|
ferredoxin 1 |
| chr8_+_91464229 | 0.07 |
ENSRNOT00000013249
|
Bckdhb
|
branched chain keto acid dehydrogenase E1 subunit beta |
| chr3_+_103945329 | 0.07 |
ENSRNOT00000008364
|
Emc7
|
ER membrane protein complex subunit 7 |
| chr14_+_88543630 | 0.07 |
ENSRNOT00000060515
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr9_-_10441834 | 0.07 |
ENSRNOT00000043704
|
Rpl36
|
ribosomal protein L36 |
| chr15_+_38069320 | 0.07 |
ENSRNOT00000014397
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr11_+_86421106 | 0.07 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr10_+_91126689 | 0.07 |
ENSRNOT00000004046
|
Nmt1
|
N-myristoyltransferase 1 |
| chr6_+_76174452 | 0.07 |
ENSRNOT00000084062
|
Psma6
|
proteasome subunit alpha 6 |
| chr4_-_77347011 | 0.07 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr8_-_40078165 | 0.07 |
ENSRNOT00000014866
|
Spa17
|
sperm autoantigenic protein 17 |
| chr10_-_47630799 | 0.07 |
ENSRNOT00000057907
|
Slc47a2
|
solute carrier family 47 member 2 |
| chr10_+_55138633 | 0.07 |
ENSRNOT00000057223
|
Mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr6_-_135112775 | 0.07 |
ENSRNOT00000086310
|
LOC103692716
|
heat shock protein HSP 90-alpha |
| chr19_-_37819789 | 0.07 |
ENSRNOT00000036702
|
Cenpt
|
centromere protein T |
| chr16_+_56247659 | 0.07 |
ENSRNOT00000017452
|
Tusc3
|
tumor suppressor candidate 3 |
| chr1_+_122981755 | 0.07 |
ENSRNOT00000013468
|
Ndn
|
necdin, MAGE family member |
| chr7_+_126736732 | 0.06 |
ENSRNOT00000022012
|
Gtse1
|
G-2 and S-phase expressed 1 |
| chr5_+_64789456 | 0.06 |
ENSRNOT00000009584
|
Zfp189
|
zinc finger protein 189 |
| chr6_-_93562314 | 0.06 |
ENSRNOT00000010871
ENSRNOT00000088790 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr6_+_26836216 | 0.06 |
ENSRNOT00000077903
ENSRNOT00000011373 |
Ost4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr7_+_27221152 | 0.06 |
ENSRNOT00000031297
|
LOC691921
|
hypothetical protein LOC691921 |
| chr1_-_220746224 | 0.06 |
ENSRNOT00000027734
|
Banf1
|
barrier to autointegration factor 1 |
| chr19_-_37370429 | 0.06 |
ENSRNOT00000022497
|
Kctd19
|
potassium channel tetramerization domain containing 19 |
| chr17_-_13593423 | 0.06 |
ENSRNOT00000019234
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr7_-_94774569 | 0.06 |
ENSRNOT00000036399
|
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr5_+_116973159 | 0.06 |
ENSRNOT00000046475
|
LOC100912024
|
uncharacterized LOC100912024 |
| chr3_-_138462063 | 0.06 |
ENSRNOT00000065553
|
Ovol2
|
ovo-like zinc finger 2 |
| chr19_+_37282018 | 0.06 |
ENSRNOT00000021723
|
Tmem208
|
transmembrane protein 208 |
| chrY_+_914045 | 0.06 |
ENSRNOT00000088593
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr10_-_104163634 | 0.06 |
ENSRNOT00000005198
|
Mif4gd
|
MIF4G domain containing |
| chr5_-_78267063 | 0.06 |
ENSRNOT00000040894
ENSRNOT00000087648 |
Cdc26
|
cell division cycle 26 |
| chr16_-_68968248 | 0.06 |
ENSRNOT00000016885
|
Eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr11_+_47243342 | 0.06 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr16_+_7292096 | 0.06 |
ENSRNOT00000025606
|
Tnnc1
|
troponin C1, slow skeletal and cardiac type |
| chr3_+_7884822 | 0.06 |
ENSRNOT00000019157
|
Med27
|
mediator complex subunit 27 |
| chr16_+_6035926 | 0.06 |
ENSRNOT00000020284
|
Selenok
|
selenoprotein K |
| chr19_+_24747178 | 0.06 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr20_-_10968432 | 0.06 |
ENSRNOT00000001593
|
Cstb
|
cystatin B |
| chr7_-_119158173 | 0.06 |
ENSRNOT00000067483
ENSRNOT00000078528 |
Txn2
|
thioredoxin 2 |
| chr9_+_100076958 | 0.06 |
ENSRNOT00000084297
|
Dusp28
|
dual specificity phosphatase 28 |
| chr2_-_188413219 | 0.06 |
ENSRNOT00000065065
|
Fdps
|
farnesyl diphosphate synthase |
| chrX_-_124766044 | 0.06 |
ENSRNOT00000000177
|
Lamp2
|
lysosomal-associated membrane protein 2 |
| chr2_+_192253629 | 0.06 |
ENSRNOT00000016364
|
Pglyrp3
|
peptidoglycan recognition protein 3 |
| chr9_+_64898459 | 0.06 |
ENSRNOT00000029526
|
Sgo2
|
shugoshin 2 |
| chr15_-_70399924 | 0.06 |
ENSRNOT00000087940
|
Diaph3
|
diaphanous-related formin 3 |
| chr11_-_34598275 | 0.06 |
ENSRNOT00000077233
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr11_-_31805728 | 0.06 |
ENSRNOT00000032162
|
Gart
|
phosphoribosylglycinamide formyltransferase |
| chrX_-_80354604 | 0.05 |
ENSRNOT00000049790
|
RGD1562039
|
similar to MGC82337 protein |
| chr8_+_117906014 | 0.05 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chrX_+_128882366 | 0.05 |
ENSRNOT00000091479
|
Tex13c2
|
TEX13 family member C2 |
| chr15_-_41691518 | 0.05 |
ENSRNOT00000020036
|
Ebpl
|
emopamil binding protein-like |
| chr5_+_151413382 | 0.05 |
ENSRNOT00000012626
|
Cd164l2
|
CD164 molecule like 2 |
| chr19_-_15733412 | 0.05 |
ENSRNOT00000014831
|
Irx6
|
iroquois homeobox 6 |
| chr9_+_84689150 | 0.05 |
ENSRNOT00000072242
|
Kcne4
|
potassium voltage-gated channel subfamily E regulatory subunit 4 |
| chr19_+_9024451 | 0.05 |
ENSRNOT00000073744
|
LOC100910721
|
60S ribosomal protein L26-like |
| chr7_-_10068532 | 0.05 |
ENSRNOT00000011102
|
AABR07055789.1
|
|
| chr11_-_38274217 | 0.05 |
ENSRNOT00000002206
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr4_-_51598771 | 0.05 |
ENSRNOT00000008325
|
Ndufa5
|
NADH:ubiquinone oxidoreductase subunit A5 |
| chr6_-_43363335 | 0.05 |
ENSRNOT00000092192
|
Itgb1bp1
|
integrin subunit beta 1 binding protein 1 |
| chr9_+_67130259 | 0.05 |
ENSRNOT00000023796
|
Cyp20a1
|
cytochrome P450, family 20, subfamily a, polypeptide 1 |
| chr8_-_49109981 | 0.05 |
ENSRNOT00000019933
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr7_-_123193761 | 0.05 |
ENSRNOT00000007164
|
Pmm1
|
phosphomannomutase 1 |
| chr13_-_110257367 | 0.05 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr4_-_165026414 | 0.05 |
ENSRNOT00000071421
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr17_-_89780691 | 0.05 |
ENSRNOT00000091830
|
LOC108348042
|
ankyrin repeat domain-containing protein 7-like |
| chr2_+_202200797 | 0.05 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr10_-_10808585 | 0.05 |
ENSRNOT00000036954
|
LOC360479
|
similar to hypothetical protein |
| chr10_-_88266210 | 0.05 |
ENSRNOT00000090702
ENSRNOT00000020603 |
Hap1
|
huntingtin-associated protein 1 |
| chr4_-_183417667 | 0.05 |
ENSRNOT00000089160
|
Fam60a
|
family with sequence similarity 60, member A |
| chr3_-_11317049 | 0.05 |
ENSRNOT00000030558
|
Swi5
|
SWI5 homologous recombination repair protein |
| chr3_-_123997277 | 0.05 |
ENSRNOT00000058238
|
RGD1564425
|
similar to Proteasome subunit beta type 3 (Proteasome theta chain) |
| chr4_-_101393329 | 0.05 |
ENSRNOT00000007636
|
RGD1562515
|
similar to RIKEN cDNA 4931417E11 |
| chr12_+_38490230 | 0.05 |
ENSRNOT00000047861
|
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr1_+_102915191 | 0.05 |
ENSRNOT00000017851
|
Ldhc
|
lactate dehydrogenase C |
| chr10_+_55924938 | 0.05 |
ENSRNOT00000087003
ENSRNOT00000057079 |
Trappc1
|
trafficking protein particle complex 1 |
| chr1_-_219144610 | 0.05 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr20_+_4967194 | 0.05 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_+_11814071 | 0.05 |
ENSRNOT00000039149
|
Tmem130
|
transmembrane protein 130 |
| chr4_+_171730635 | 0.05 |
ENSRNOT00000009923
|
Strap
|
serine/threonine kinase receptor associated protein |
| chr3_-_164366684 | 0.05 |
ENSRNOT00000035048
|
LOC679539
|
similar to ubiquitin-conjugating enzyme E2 variant 1 |
| chr6_+_42568220 | 0.05 |
ENSRNOT00000074829
|
Pdia6
|
protein disulfide isomerase family A, member 6 |
| chr7_-_12918173 | 0.05 |
ENSRNOT00000011010
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr5_+_128450680 | 0.05 |
ENSRNOT00000010700
|
Txndc12
|
thioredoxin domain containing 12 |
| chr7_-_140617721 | 0.04 |
ENSRNOT00000081355
|
Tuba1b
|
tubulin, alpha 1B |
| chr4_+_38240728 | 0.04 |
ENSRNOT00000047029
|
Phf14
|
PHD finger protein 14 |
| chr11_+_64882288 | 0.04 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr3_+_13838304 | 0.04 |
ENSRNOT00000025067
|
Hspa5
|
heat shock protein family A member 5 |
| chr19_+_24846938 | 0.04 |
ENSRNOT00000045974
|
Ddx39a
|
DExD-box helicase 39A |
| chr14_+_33354807 | 0.04 |
ENSRNOT00000049774
|
Hopx
|
HOP homeobox |
| chr7_+_143924662 | 0.04 |
ENSRNOT00000017794
|
Pfdn5
|
prefoldin subunit 5 |
| chr15_-_28517335 | 0.04 |
ENSRNOT00000060514
ENSRNOT00000090378 |
LOC100911576
|
heterogeneous nuclear ribonucleoproteins C1/C2-like |
| chr19_-_869490 | 0.04 |
ENSRNOT00000080433
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr9_-_73871888 | 0.04 |
ENSRNOT00000017686
|
Acadl
|
acyl-CoA dehydrogenase, long chain |
| chr7_+_123296183 | 0.04 |
ENSRNOT00000048614
|
LOC100362109
|
hypothetical protein LOC100362109 |
| chr7_+_72599479 | 0.04 |
ENSRNOT00000018963
|
AABR07057460.1
|
|
| chr4_+_27175243 | 0.04 |
ENSRNOT00000009985
|
Cyp51
|
cytochrome P450, family 51 |
| chr3_-_2181962 | 0.04 |
ENSRNOT00000044802
|
Mrpl41
|
mitochondrial ribosomal protein L41 |
| chr4_-_151516894 | 0.04 |
ENSRNOT00000011044
|
Wnt5b
|
wingless-type MMTV integration site family, member 5B |
| chr17_+_30965942 | 0.04 |
ENSRNOT00000060143
|
Pxdc1
|
PX domain containing 1 |
| chr12_+_10255416 | 0.04 |
ENSRNOT00000092720
|
Gpr12
|
G protein-coupled receptor 12 |
| chr4_-_164453171 | 0.04 |
ENSRNOT00000077539
ENSRNOT00000083610 ENSRNOT00000079975 |
Ly49s6
|
Ly49 stimulatory receptor 6 |
| chr16_+_54765325 | 0.04 |
ENSRNOT00000065327
ENSRNOT00000086899 |
Mtmr7
|
myotubularin related protein 7 |
| chr17_-_44758170 | 0.04 |
ENSRNOT00000091176
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr3_+_137137980 | 0.04 |
ENSRNOT00000006900
|
Snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr8_+_22021213 | 0.04 |
ENSRNOT00000049706
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr2_+_46980976 | 0.04 |
ENSRNOT00000083668
ENSRNOT00000082990 |
Mocs2
|
molybdenum cofactor synthesis 2 |
| chr7_+_116357175 | 0.04 |
ENSRNOT00000076790
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr1_+_114046478 | 0.04 |
ENSRNOT00000032254
|
Siglech
|
sialic acid binding Ig-like lectin H |
| chr14_-_84662143 | 0.04 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr15_+_52220578 | 0.04 |
ENSRNOT00000015104
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr11_-_64583994 | 0.04 |
ENSRNOT00000004289
|
B4galt4
|
beta-1,4-galactosyltransferase 4 |
| chr14_-_18862407 | 0.04 |
ENSRNOT00000003823
|
Cxcl6
|
C-X-C motif chemokine ligand 6 |
| chr6_-_104289668 | 0.04 |
ENSRNOT00000085646
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr10_+_94466523 | 0.04 |
ENSRNOT00000014867
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr14_+_33010300 | 0.04 |
ENSRNOT00000002809
|
Igfbp7
|
insulin-like growth factor binding protein 7 |
| chr9_+_16543688 | 0.04 |
ENSRNOT00000021868
|
Cnpy3
|
canopy FGF signaling regulator 3 |
| chr3_-_81886545 | 0.04 |
ENSRNOT00000057007
|
LOC102547078
|
myb-related transcription factor, partner of profilin-like |
| chr9_+_100156154 | 0.04 |
ENSRNOT00000005935
|
Aqp12a
|
aquaporin 12A |
| chr10_+_11338306 | 0.04 |
ENSRNOT00000033695
|
LOC100910875
|
protein FAM100A-like |
| chr8_-_59849427 | 0.04 |
ENSRNOT00000020332
ENSRNOT00000083900 |
Nrg4
|
neuregulin 4 |
| chr6_+_72891725 | 0.04 |
ENSRNOT00000038074
|
Nubpl
|
nucleotide binding protein-like |
| chr6_+_129835919 | 0.04 |
ENSRNOT00000036035
|
Vrk1
|
vaccinia related kinase 1 |
| chr6_+_108796182 | 0.04 |
ENSRNOT00000006297
|
Fcf1
|
FCF1 rRNA-processing protein |
| chr19_-_10681145 | 0.04 |
ENSRNOT00000022167
|
Ccl22
|
C-C motif chemokine ligand 22 |
| chr7_-_30105132 | 0.04 |
ENSRNOT00000091227
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr8_-_27852996 | 0.04 |
ENSRNOT00000037790
|
Glb1l2
|
galactosidase, beta 1-like 2 |
| chrX_+_31054394 | 0.04 |
ENSRNOT00000037439
|
LOC688526
|
similar to ribonucleic acid binding protein S1 |
| chr6_-_127337791 | 0.04 |
ENSRNOT00000032015
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr1_+_220746387 | 0.04 |
ENSRNOT00000027753
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr10_+_10555337 | 0.04 |
ENSRNOT00000032495
|
RGD1565166
|
similar to MGC45438 protein |
| chr17_+_9289189 | 0.04 |
ENSRNOT00000015632
|
H2afy
|
H2A histone family, member Y |
| chr1_+_79030291 | 0.04 |
ENSRNOT00000037086
ENSRNOT00000088925 |
Cgm4
|
carcinoembryonic antigen gene family 4 |
| chrM_+_7758 | 0.04 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.2 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.1 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.2 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0045901 | regulation of translational termination(GO:0006449) positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0000105 | histidine biosynthetic process(GO:0000105) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.0 | GO:0019516 | lactate oxidation(GO:0019516) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.0 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.0 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.0 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0015820 | leucine transport(GO:0015820) tryptophan transport(GO:0015827) |
| 0.0 | 0.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:0035983 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.0 | 0.1 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) lysosomal matrix(GO:1990836) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.0 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0032551 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.0 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |