Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
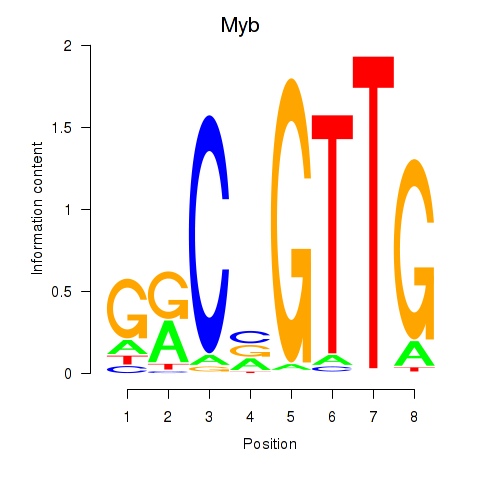
Results for Myb
Z-value: 0.29
Transcription factors associated with Myb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Myb
|
ENSRNOG00000055858 | MYB proto-oncogene, transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Myb | rn6_v1_chr1_-_16687817_16687817 | 0.34 | 5.7e-01 | Click! |
Activity profile of Myb motif
Sorted Z-values of Myb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_56531483 | 0.15 |
ENSRNOT00000022343
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr12_+_24367199 | 0.12 |
ENSRNOT00000001971
|
Fkbp6
|
FK506 binding protein 6 |
| chr4_+_85009350 | 0.11 |
ENSRNOT00000072271
|
Znrf2
|
zinc and ring finger 2 |
| chr13_-_110257367 | 0.08 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr7_-_73216251 | 0.08 |
ENSRNOT00000057587
|
LOC100362027
|
ribosomal protein L30-like |
| chr2_-_187668677 | 0.07 |
ENSRNOT00000056898
ENSRNOT00000092563 |
Tsacc
|
TSSK6 activating co-chaperone |
| chr20_-_28873363 | 0.07 |
ENSRNOT00000071064
|
Snrpd2l
|
small nuclear ribonucleoprotein D2-like |
| chr3_-_2490392 | 0.07 |
ENSRNOT00000014993
|
Ssna1
|
SS nuclear autoantigen 1 |
| chr16_+_19001061 | 0.07 |
ENSRNOT00000016355
|
Med26
|
mediator complex subunit 26 |
| chr11_+_1896209 | 0.06 |
ENSRNOT00000000909
|
Cggbp1
|
CGG triplet repeat binding protein 1 |
| chr1_-_162713610 | 0.06 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr15_-_34352673 | 0.06 |
ENSRNOT00000064916
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr3_-_138462063 | 0.06 |
ENSRNOT00000065553
|
Ovol2
|
ovo-like zinc finger 2 |
| chr10_+_57251253 | 0.06 |
ENSRNOT00000043171
|
LOC108348287
|
60S ribosomal protein L36a |
| chr8_-_64572850 | 0.06 |
ENSRNOT00000015415
|
Senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr2_-_116161998 | 0.06 |
ENSRNOT00000012560
|
Gpr160
|
G protein-coupled receptor 160 |
| chr16_+_74292438 | 0.06 |
ENSRNOT00000026197
|
Vdac3
|
voltage-dependent anion channel 3 |
| chr7_+_2479311 | 0.06 |
ENSRNOT00000003787
|
Ptges3
|
prostaglandin E synthase 3 |
| chr5_-_101588598 | 0.05 |
ENSRNOT00000082239
|
Psip1
|
PC4 and SFRS1 interacting protein 1 |
| chr2_-_84531192 | 0.05 |
ENSRNOT00000065312
ENSRNOT00000090540 |
Ropn1l
|
rhophilin associated tail protein 1-like |
| chr7_+_3320103 | 0.05 |
ENSRNOT00000090381
|
Cd63
|
Cd63 molecule |
| chr1_-_146289465 | 0.05 |
ENSRNOT00000017362
|
Abhd17c
|
abhydrolase domain containing 17C |
| chr16_-_7082193 | 0.05 |
ENSRNOT00000024447
|
Spcs1
|
signal peptidase complex subunit 1 |
| chr8_+_79054237 | 0.05 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr1_+_144070754 | 0.05 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr6_+_26517840 | 0.05 |
ENSRNOT00000031842
|
Ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr3_-_2727616 | 0.05 |
ENSRNOT00000061904
|
C8g
|
complement C8 gamma chain |
| chr2_-_188252725 | 0.05 |
ENSRNOT00000027608
|
Dap3
|
death associated protein 3 |
| chr6_+_51662224 | 0.05 |
ENSRNOT00000060006
|
Ccdc71l
|
coiled-coil domain containing 71-like |
| chr2_+_206392200 | 0.05 |
ENSRNOT00000026631
|
Rsbn1
|
round spermatid basic protein 1 |
| chr1_-_277181345 | 0.05 |
ENSRNOT00000038017
ENSRNOT00000038038 |
Nrap
|
nebulin-related anchoring protein |
| chr11_-_31892531 | 0.04 |
ENSRNOT00000033745
|
Cryzl1
|
crystallin zeta like 1 |
| chr15_-_26175645 | 0.04 |
ENSRNOT00000020175
|
Slc35f4
|
solute carrier family 35, member F4 |
| chr5_-_137265015 | 0.04 |
ENSRNOT00000036151
|
Cdc20
|
cell division cycle 20 |
| chr19_+_24846938 | 0.04 |
ENSRNOT00000045974
|
Ddx39a
|
DExD-box helicase 39A |
| chr2_+_252263386 | 0.04 |
ENSRNOT00000092913
ENSRNOT00000084034 ENSRNOT00000041186 ENSRNOT00000092931 |
Ssx2ip
|
SSX family member 2 interacting protein |
| chr10_+_92628356 | 0.04 |
ENSRNOT00000072480
|
Myl4
|
myosin, light chain 4 |
| chr14_-_84662143 | 0.04 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr15_+_38069320 | 0.04 |
ENSRNOT00000014397
|
Sap18
|
Sin3-associated polypeptide 18 |
| chr8_-_117353672 | 0.04 |
ENSRNOT00000027262
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr2_-_189899325 | 0.04 |
ENSRNOT00000017561
|
Chtop
|
chromatin target of PRMT1 |
| chr18_+_63394900 | 0.04 |
ENSRNOT00000024126
|
Psmg2
|
proteasome assembly chaperone 2 |
| chr11_-_57993548 | 0.04 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr19_+_43217161 | 0.04 |
ENSRNOT00000082566
|
Ddx19a
|
DEAD-box helicase 19A |
| chr1_+_219077771 | 0.04 |
ENSRNOT00000022824
|
Chka
|
choline kinase alpha |
| chr4_+_6827429 | 0.04 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr9_-_50868238 | 0.04 |
ENSRNOT00000015600
|
Tex30
|
testis expressed 30 |
| chr2_+_127538659 | 0.04 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr6_-_128727374 | 0.04 |
ENSRNOT00000082152
|
Syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr10_+_40812858 | 0.04 |
ENSRNOT00000018149
|
G3bp1
|
G3BP stress granule assembly factor 1 |
| chr14_-_115352562 | 0.03 |
ENSRNOT00000050321
|
Erlec1
|
endoplasmic reticulum lectin 1 |
| chr7_+_64987859 | 0.03 |
ENSRNOT00000005694
|
Tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chrX_-_72034099 | 0.03 |
ENSRNOT00000004310
|
Ercc6l
|
ERCC excision repair 6 like, spindle assembly checkpoint helicase |
| chr3_+_66193059 | 0.03 |
ENSRNOT00000006880
|
Itga4
|
integrin subunit alpha 4 |
| chr5_-_172488822 | 0.03 |
ENSRNOT00000019620
|
Rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr10_+_55138633 | 0.03 |
ENSRNOT00000057223
|
Mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr8_+_117812099 | 0.03 |
ENSRNOT00000077228
|
Ccdc51
|
coiled-coil domain containing 51 |
| chr10_-_84976170 | 0.03 |
ENSRNOT00000013542
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr4_-_77347011 | 0.03 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr2_-_189899103 | 0.03 |
ENSRNOT00000010455
|
Chtop
|
chromatin target of PRMT1 |
| chr2_-_188252551 | 0.03 |
ENSRNOT00000088327
|
Dap3
|
death associated protein 3 |
| chr10_-_89199736 | 0.03 |
ENSRNOT00000027782
|
Coa3
|
cytochrome C oxidase assembly factor 3 |
| chr5_+_150725654 | 0.03 |
ENSRNOT00000089852
ENSRNOT00000017740 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr11_-_1819094 | 0.03 |
ENSRNOT00000000911
|
MGC95208
|
similar to 4930453N24Rik protein |
| chr3_+_9643047 | 0.03 |
ENSRNOT00000035805
|
Ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr8_+_64573358 | 0.03 |
ENSRNOT00000083558
|
Myo9a
|
myosin IXA |
| chr2_+_208323882 | 0.03 |
ENSRNOT00000085178
|
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr1_-_260638816 | 0.03 |
ENSRNOT00000065632
ENSRNOT00000017938 ENSRNOT00000077962 |
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr7_-_117576737 | 0.03 |
ENSRNOT00000039795
|
Dgat1
|
diacylglycerol O-acyltransferase 1 |
| chr13_-_47916877 | 0.03 |
ENSRNOT00000006502
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr1_-_279277339 | 0.03 |
ENSRNOT00000023667
|
Gfra1
|
GDNF family receptor alpha 1 |
| chr15_-_41890716 | 0.03 |
ENSRNOT00000020419
|
Spryd7
|
SPRY domain containing 7 |
| chr7_+_40316639 | 0.03 |
ENSRNOT00000080150
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr5_+_159606647 | 0.03 |
ENSRNOT00000051693
|
Rps21-ps1
|
ribosomal protein S21, pseudogene 1 |
| chr1_+_102915191 | 0.03 |
ENSRNOT00000017851
|
Ldhc
|
lactate dehydrogenase C |
| chr9_+_64898459 | 0.03 |
ENSRNOT00000029526
|
Sgo2
|
shugoshin 2 |
| chr5_-_147953093 | 0.03 |
ENSRNOT00000075270
|
Khdrbs1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chr9_+_40817654 | 0.03 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chr1_+_51619875 | 0.03 |
ENSRNOT00000023319
|
Pabpc6
|
poly(A) binding protein, cytoplasmic 6 |
| chr9_+_19917603 | 0.03 |
ENSRNOT00000038239
|
Tdrd6
|
tudor domain containing 6 |
| chr1_+_255040426 | 0.03 |
ENSRNOT00000092151
|
Pcgf5
|
polycomb group ring finger 5 |
| chr3_+_153398130 | 0.03 |
ENSRNOT00000068135
|
Rpn2
|
ribophorin II |
| chr8_-_115626380 | 0.03 |
ENSRNOT00000019107
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr2_+_52301798 | 0.02 |
ENSRNOT00000091387
|
Paip1
|
poly(A) binding protein interacting protein 1 |
| chr13_+_97702097 | 0.02 |
ENSRNOT00000057787
|
LOC103689999
|
saccharopine dehydrogenase-like oxidoreductase |
| chr6_-_43493816 | 0.02 |
ENSRNOT00000077786
|
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr1_-_198298076 | 0.02 |
ENSRNOT00000027033
|
Ino80e
|
INO80 complex subunit E |
| chr15_+_34352914 | 0.02 |
ENSRNOT00000067150
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr9_-_14551519 | 0.02 |
ENSRNOT00000067623
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr12_+_2461502 | 0.02 |
ENSRNOT00000001415
|
Elavl1
|
ELAV like RNA binding protein 1 |
| chr5_-_13097341 | 0.02 |
ENSRNOT00000061884
|
Rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr5_-_24255606 | 0.02 |
ENSRNOT00000037483
|
Plekhf2
|
pleckstrin homology and FYVE domain containing 2 |
| chr17_-_43675934 | 0.02 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr12_+_38490230 | 0.02 |
ENSRNOT00000047861
|
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr1_-_199037267 | 0.02 |
ENSRNOT00000078779
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr1_+_265298868 | 0.02 |
ENSRNOT00000023278
|
Dpcd
|
deleted in primary ciliary dyskinesia |
| chr4_-_165629996 | 0.02 |
ENSRNOT00000068185
ENSRNOT00000007427 |
Ybx3
|
Y box binding protein 3 |
| chr6_-_114488880 | 0.02 |
ENSRNOT00000087560
|
AC118957.1
|
|
| chr13_-_85443727 | 0.02 |
ENSRNOT00000090668
ENSRNOT00000076125 |
Uck2
|
uridine-cytidine kinase 2 |
| chr14_+_85311089 | 0.02 |
ENSRNOT00000080541
|
Rasl10a
|
RAS-like, family 10, member A |
| chr10_-_89699836 | 0.02 |
ENSRNOT00000084311
|
Etv4
|
ets variant 4 |
| chr1_-_31528083 | 0.02 |
ENSRNOT00000061041
|
Rpl26-ps2
|
ribosomal protein L26, pseudogene 2 |
| chr19_+_14392423 | 0.02 |
ENSRNOT00000018880
|
Tom1
|
target of myb1 membrane trafficking protein |
| chr20_-_11737050 | 0.02 |
ENSRNOT00000001637
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chr17_-_80577027 | 0.02 |
ENSRNOT00000057827
|
Rsu1
|
Ras suppressor protein 1 |
| chr18_-_40518988 | 0.02 |
ENSRNOT00000004784
|
Fem1c
|
fem-1 homolog C |
| chr5_-_134871775 | 0.02 |
ENSRNOT00000015667
|
LOC100911581
|
fatty-acid amide hydrolase 1-like |
| chr19_+_37127508 | 0.02 |
ENSRNOT00000019656
|
Cbfb
|
core-binding factor, beta subunit |
| chr15_+_32763067 | 0.02 |
ENSRNOT00000011998
|
AABR07017902.1
|
|
| chr2_+_181331464 | 0.02 |
ENSRNOT00000017448
|
Map9
|
microtubule-associated protein 9 |
| chrX_-_139464798 | 0.02 |
ENSRNOT00000003282
|
Gpc4
|
glypican 4 |
| chr18_+_29386809 | 0.02 |
ENSRNOT00000082079
ENSRNOT00000024825 |
Eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr16_-_7336335 | 0.02 |
ENSRNOT00000044991
|
Phf7
|
PHD finger protein 7 |
| chr7_-_12640232 | 0.02 |
ENSRNOT00000014981
|
Elane
|
elastase, neutrophil expressed |
| chr1_-_263845762 | 0.02 |
ENSRNOT00000017268
|
Erlin1
|
ER lipid raft associated 1 |
| chr16_+_2379480 | 0.02 |
ENSRNOT00000079215
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr13_-_95943761 | 0.02 |
ENSRNOT00000005961
|
Adss
|
adenylosuccinate synthase |
| chr5_+_169357517 | 0.02 |
ENSRNOT00000000079
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr8_-_59313521 | 0.02 |
ENSRNOT00000017239
ENSRNOT00000090439 |
Wdr61
|
WD repeat domain 61 |
| chr4_+_157348020 | 0.02 |
ENSRNOT00000020803
|
Cdca3
|
cell division cycle associated 3 |
| chr16_-_21338771 | 0.02 |
ENSRNOT00000014265
|
Pbx4
|
PBX homeobox 4 |
| chr6_-_135259603 | 0.02 |
ENSRNOT00000010529
|
Mok
|
MOK protein kinase |
| chr6_-_109103999 | 0.02 |
ENSRNOT00000009086
ENSRNOT00000080009 |
Acyp1
|
acylphosphatase 1 |
| chr16_+_7336685 | 0.02 |
ENSRNOT00000025853
|
Bap1
|
Brca1 associated protein 1 |
| chr10_+_56546710 | 0.02 |
ENSRNOT00000023003
|
Ybx2
|
Y box binding protein 2 |
| chr15_+_52265557 | 0.02 |
ENSRNOT00000015969
|
Nudt18
|
nudix hydrolase 18 |
| chr8_+_117170620 | 0.02 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr15_-_28044210 | 0.02 |
ENSRNOT00000033809
|
Rnase1l1
|
ribonuclease, RNase A family, 1-like 1 (pancreatic) |
| chr14_-_72122158 | 0.02 |
ENSRNOT00000064495
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr1_-_101803200 | 0.02 |
ENSRNOT00000028588
|
Cyth2
|
cytohesin 2 |
| chr6_+_136536736 | 0.02 |
ENSRNOT00000086594
|
Tdrd9
|
tudor domain containing 9 |
| chr7_-_121058029 | 0.02 |
ENSRNOT00000068033
|
Cbx6
|
chromobox 6 |
| chr1_+_257517242 | 0.02 |
ENSRNOT00000073502
ENSRNOT00000018668 ENSRNOT00000078738 ENSRNOT00000091663 ENSRNOT00000072002 |
LOC685933
|
hypothetical protein LOC685933 |
| chr18_-_79258570 | 0.02 |
ENSRNOT00000022401
|
Galr1
|
galanin receptor 1 |
| chr19_+_24545318 | 0.02 |
ENSRNOT00000005071
|
Clgn
|
calmegin |
| chr11_-_88038518 | 0.02 |
ENSRNOT00000051991
|
Rimbp3
|
RIMS binding protein 3 |
| chr4_-_170129407 | 0.02 |
ENSRNOT00000075717
|
LOC100912609
|
uncharacterized LOC100912609 |
| chr7_-_75421874 | 0.02 |
ENSRNOT00000012775
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr6_-_109692218 | 0.02 |
ENSRNOT00000012327
|
RGD1310769
|
similar to HSPC288 |
| chr16_-_19376776 | 0.02 |
ENSRNOT00000020748
|
Rab8a
|
RAB8A, member RAS oncogene family |
| chr1_+_79911516 | 0.02 |
ENSRNOT00000019075
|
Irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr3_+_2490518 | 0.01 |
ENSRNOT00000015099
|
Anapc2
|
anaphase promoting complex subunit 2 |
| chr3_+_92969141 | 0.01 |
ENSRNOT00000009797
|
Apip
|
APAF1 interacting protein |
| chr19_-_914880 | 0.01 |
ENSRNOT00000017127
|
Cklf
|
chemokine-like factor |
| chr6_-_23543172 | 0.01 |
ENSRNOT00000006073
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr19_+_24747178 | 0.01 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr12_+_39679688 | 0.01 |
ENSRNOT00000050398
|
Fam216a
|
family with sequence similarity 216, member A |
| chr1_+_80135391 | 0.01 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr14_+_114126943 | 0.01 |
ENSRNOT00000041638
ENSRNOT00000006443 ENSRNOT00000006957 |
Rtn4
|
reticulon 4 |
| chr7_-_25743537 | 0.01 |
ENSRNOT00000083450
|
LOC100910996
|
uncharacterized LOC100910996 |
| chr15_-_42751330 | 0.01 |
ENSRNOT00000066478
|
Adam2
|
ADAM metallopeptidase domain 2 |
| chr6_-_23542928 | 0.01 |
ENSRNOT00000083858
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr6_+_135549137 | 0.01 |
ENSRNOT00000073989
|
Rcor1
|
REST corepressor 1 |
| chr4_-_57625147 | 0.01 |
ENSRNOT00000074649
ENSRNOT00000078961 |
Ube2h
|
ubiquitin-conjugating enzyme E2H |
| chr10_+_730247 | 0.01 |
ENSRNOT00000083714
|
Fopnl
|
FGFR1OP N-terminal like |
| chr7_+_125937928 | 0.01 |
ENSRNOT00000044274
|
Fam118a
|
family with sequence similarity 118, member A |
| chr6_+_78567970 | 0.01 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr13_-_79801368 | 0.01 |
ENSRNOT00000075998
ENSRNOT00000084058 |
Suco
|
SUN domain containing ossification factor |
| chr18_-_40586260 | 0.01 |
ENSRNOT00000004942
|
Tmed7
|
transmembrane p24 trafficking protein 7 |
| chr2_-_178389608 | 0.01 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr16_+_7082450 | 0.01 |
ENSRNOT00000013291
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr8_-_117811975 | 0.01 |
ENSRNOT00000028055
|
Atrip
|
ATR interacting protein |
| chr10_-_89700283 | 0.01 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr20_+_5509059 | 0.01 |
ENSRNOT00000065349
|
Kifc1
|
kinesin family member C1 |
| chr3_-_2770620 | 0.01 |
ENSRNOT00000008253
|
Traf2
|
Tnf receptor-associated factor 2 |
| chr12_-_31323810 | 0.01 |
ENSRNOT00000001247
|
Ran
|
RAN, member RAS oncogene family |
| chr1_-_254671596 | 0.01 |
ENSRNOT00000025450
|
Htr7
|
5-hydroxytryptamine receptor 7 |
| chr11_+_60072727 | 0.01 |
ENSRNOT00000090230
|
Tagln3
|
transgelin 3 |
| chr1_-_101426852 | 0.01 |
ENSRNOT00000028217
|
Ruvbl2
|
RuvB-like AAA ATPase 2 |
| chr2_-_89310946 | 0.01 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr3_+_8982122 | 0.01 |
ENSRNOT00000024914
|
Ptpa
|
protein phosphatase 2 phosphatase activator |
| chr3_+_110618298 | 0.01 |
ENSRNOT00000012454
|
Knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr8_-_64268555 | 0.01 |
ENSRNOT00000013581
ENSRNOT00000084758 |
Arih1
|
ariadne RBR E3 ubiquitin protein ligase 1 |
| chr6_-_2886465 | 0.01 |
ENSRNOT00000038448
|
Srsf7
|
serine and arginine rich splicing factor 7 |
| chr5_+_171297850 | 0.01 |
ENSRNOT00000034284
|
Lrrc47
|
leucine rich repeat containing 47 |
| chr10_-_31493419 | 0.01 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chr1_-_165382216 | 0.01 |
ENSRNOT00000023648
|
Ppme1
|
protein phosphatase methylesterase 1 |
| chr7_+_37812831 | 0.01 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr3_-_14643897 | 0.01 |
ENSRNOT00000082008
ENSRNOT00000025983 |
Ggta1
|
glycoprotein, alpha-galactosyltransferase 1 |
| chr15_-_45927804 | 0.01 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr20_+_50652745 | 0.01 |
ENSRNOT00000000365
|
Hace1
|
HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 |
| chr10_-_85709568 | 0.01 |
ENSRNOT00000005426
|
Cwc25
|
CWC25 spliceosome-associated protein homolog |
| chr6_+_107596782 | 0.01 |
ENSRNOT00000076882
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr1_+_82169620 | 0.01 |
ENSRNOT00000088955
ENSRNOT00000068251 |
Prr19
|
proline rich 19 |
| chr19_+_39126589 | 0.01 |
ENSRNOT00000027561
|
Sntb2
|
syntrophin, beta 2 |
| chr7_-_75597276 | 0.01 |
ENSRNOT00000035628
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr3_-_93734282 | 0.01 |
ENSRNOT00000012428
|
Caprin1
|
cell cycle associated protein 1 |
| chr1_-_265298797 | 0.01 |
ENSRNOT00000023137
|
Poll
|
DNA polymerase lambda |
| chr4_-_145329878 | 0.01 |
ENSRNOT00000011827
|
Tada3
|
transcriptional adaptor 3 |
| chr2_+_188253220 | 0.01 |
ENSRNOT00000027629
|
Ash1l
|
ASH1 like histone lysine methyltransferase |
| chr10_+_90622992 | 0.01 |
ENSRNOT00000032856
|
Meioc
|
meiosis specific with coiled-coil domain |
| chr3_+_79678201 | 0.01 |
ENSRNOT00000087604
ENSRNOT00000079709 |
Mtch2
|
mitochondrial carrier 2 |
| chr18_+_59096643 | 0.01 |
ENSRNOT00000025325
|
Wdr7
|
WD repeat domain 7 |
| chr10_-_65329279 | 0.01 |
ENSRNOT00000088721
ENSRNOT00000078757 ENSRNOT00000014104 ENSRNOT00000014118 |
Flot2
|
flotillin 2 |
| chr1_+_247110216 | 0.01 |
ENSRNOT00000016000
|
Cdc37l1
|
cell division cycle 37-like 1 |
| chr3_-_161498951 | 0.01 |
ENSRNOT00000024214
|
Ncoa5
|
nuclear receptor coactivator 5 |
| chr4_-_55527092 | 0.01 |
ENSRNOT00000087407
ENSRNOT00000010223 |
Zfp800
|
zinc finger protein 800 |
| chr5_+_154205402 | 0.01 |
ENSRNOT00000064925
|
Srsf10
|
serine and arginine rich splicing factor 10 |
| chr1_+_219345918 | 0.01 |
ENSRNOT00000025018
|
Cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr1_+_23977688 | 0.01 |
ENSRNOT00000014805
|
Tbpl1
|
TATA-box binding protein like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Myb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.1 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.0 | GO:1903165 | response to polycyclic arene(GO:1903165) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |