Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
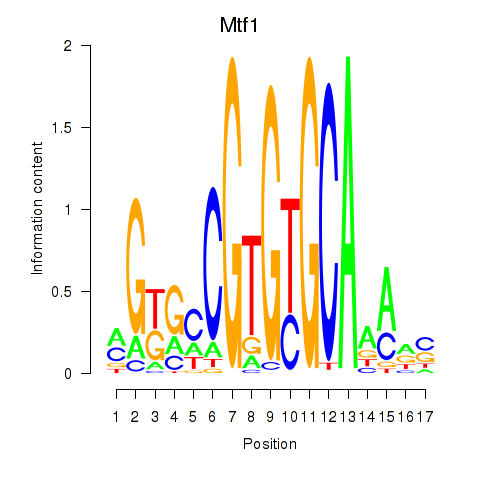
Results for Mtf1
Z-value: 0.31
Transcription factors associated with Mtf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mtf1
|
ENSRNOG00000025724 | metal-regulatory transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mtf1 | rn6_v1_chr5_+_142797366_142797366 | -0.94 | 1.9e-02 | Click! |
Activity profile of Mtf1 motif
Sorted Z-values of Mtf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_11302938 | 0.50 |
ENSRNOT00000038212
|
AC128848.1
|
|
| chr19_-_11308740 | 0.28 |
ENSRNOT00000067391
|
Mt2A
|
metallothionein 2A |
| chr4_+_170149029 | 0.12 |
ENSRNOT00000073287
|
LOC103690002
|
histone H2A.J |
| chr1_+_72882806 | 0.11 |
ENSRNOT00000024640
|
Tnni3
|
troponin I3, cardiac type |
| chr2_+_198359754 | 0.10 |
ENSRNOT00000048582
|
Hist2h2ab
|
histone cluster 2 H2A family member b |
| chr13_+_52553775 | 0.10 |
ENSRNOT00000011991
|
Csrp1
|
cysteine and glycine-rich protein 1 |
| chr19_-_38189523 | 0.08 |
ENSRNOT00000027200
|
Slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr4_-_123494742 | 0.08 |
ENSRNOT00000073268
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr4_-_129430251 | 0.08 |
ENSRNOT00000067881
|
Fam19a4
|
family with sequence similarity 19 member A4, C-C motif chemokine like |
| chr13_+_75059927 | 0.07 |
ENSRNOT00000080801
|
LOC680254
|
hypothetical protein LOC680254 |
| chr3_+_124088157 | 0.07 |
ENSRNOT00000028876
|
Smox
|
spermine oxidase |
| chr1_-_77808462 | 0.07 |
ENSRNOT00000078917
ENSRNOT00000080422 |
LOC103689961
|
selenoprotein W-like |
| chr1_-_77535681 | 0.07 |
ENSRNOT00000018559
|
Selenow
|
selenoprotein W |
| chr20_-_5806097 | 0.06 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chrX_+_15035569 | 0.05 |
ENSRNOT00000006491
ENSRNOT00000078392 |
Porcn
|
porcupine homolog (Drosophila) |
| chr2_-_45077219 | 0.05 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr7_-_116963281 | 0.05 |
ENSRNOT00000082058
ENSRNOT00000012306 |
Tsta3
|
tissue specific transplantation antigen P35B |
| chr1_+_12823363 | 0.05 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr19_+_25983169 | 0.05 |
ENSRNOT00000004404
|
Syce2
|
synaptonemal complex central element protein 2 |
| chr5_-_106758032 | 0.05 |
ENSRNOT00000033651
|
Hacd4
|
3-hydroxyacyl-CoA dehydratase 4 |
| chr14_-_86706626 | 0.04 |
ENSRNOT00000082893
|
H2afv
|
H2A histone family, member V |
| chr13_+_97702097 | 0.04 |
ENSRNOT00000057787
|
LOC103689999
|
saccharopine dehydrogenase-like oxidoreductase |
| chr4_-_119756765 | 0.04 |
ENSRNOT00000013884
|
Cnbp
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr19_-_52282877 | 0.04 |
ENSRNOT00000021271
|
Kcng4
|
potassium voltage-gated channel modifier subfamily G member 4 |
| chr3_+_175548174 | 0.04 |
ENSRNOT00000091941
|
Adrm1
|
adhesion regulating molecule 1 |
| chr9_-_119321500 | 0.04 |
ENSRNOT00000048125
|
Myl12b
|
myosin light chain 12B |
| chr9_+_16543688 | 0.04 |
ENSRNOT00000021868
|
Cnpy3
|
canopy FGF signaling regulator 3 |
| chr12_-_11265865 | 0.04 |
ENSRNOT00000001315
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr6_-_55647665 | 0.03 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr20_-_5805627 | 0.03 |
ENSRNOT00000085996
|
Clps
|
colipase |
| chr4_-_115453659 | 0.03 |
ENSRNOT00000065847
|
Tex261
|
testis expressed 261 |
| chr5_+_152559577 | 0.03 |
ENSRNOT00000082245
|
Slc30a2
|
solute carrier family 30 member 2 |
| chr1_-_81377976 | 0.03 |
ENSRNOT00000075189
|
Srrm5
|
serine/arginine repetitive matrix 5 |
| chr1_+_189549960 | 0.03 |
ENSRNOT00000019654
|
Exnef
|
exonuclease NEF-sp |
| chr16_-_40555576 | 0.03 |
ENSRNOT00000015529
|
Vegfc
|
vascular endothelial growth factor C |
| chr19_+_55300395 | 0.03 |
ENSRNOT00000092169
|
Ctu2
|
cytosolic thiouridylase subunit 2 |
| chrX_+_74497262 | 0.03 |
ENSRNOT00000003899
|
Zcchc13
|
zinc finger CCHC-type containing 13 |
| chr15_+_18540913 | 0.03 |
ENSRNOT00000010545
|
Pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr20_+_18492169 | 0.03 |
ENSRNOT00000046536
|
LOC103690821
|
60S ribosomal protein L39 |
| chr7_-_141307233 | 0.03 |
ENSRNOT00000071885
|
Racgap1
|
Rac GTPase-activating protein 1 |
| chr7_-_117679219 | 0.03 |
ENSRNOT00000071522
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr15_-_37410848 | 0.03 |
ENSRNOT00000081757
|
Gjb6
|
gap junction protein, beta 6 |
| chrX_+_28435507 | 0.03 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr9_+_93030714 | 0.03 |
ENSRNOT00000023581
|
Spata3
|
spermatogenesis associated 3 |
| chr1_+_217018916 | 0.02 |
ENSRNOT00000028195
ENSRNOT00000078979 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr10_+_29165577 | 0.02 |
ENSRNOT00000078415
|
Ccnjl
|
cyclin J-like |
| chr6_+_127400585 | 0.02 |
ENSRNOT00000087429
|
Ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr16_+_47368768 | 0.02 |
ENSRNOT00000017966
|
Wwc2
|
WW and C2 domain containing 2 |
| chrX_-_16526028 | 0.02 |
ENSRNOT00000093647
|
Dgkk
|
diacylglycerol kinase kappa |
| chr19_+_56179111 | 0.02 |
ENSRNOT00000046155
|
Tcf25
|
transcription factor 25 |
| chr3_-_171166454 | 0.02 |
ENSRNOT00000034131
|
Ctcfl
|
CCCTC-binding factor like |
| chr8_-_71784551 | 0.02 |
ENSRNOT00000022960
|
Snx1
|
sorting nexin 1 |
| chr6_-_132747828 | 0.02 |
ENSRNOT00000005753
|
Slc25a29
|
solute carrier family 25 member 29 |
| chr1_-_213921208 | 0.02 |
ENSRNOT00000044393
|
Ano9
|
anoctamin 9 |
| chr20_-_11737050 | 0.02 |
ENSRNOT00000001637
|
Sumo3
|
small ubiquitin-like modifier 3 |
| chr4_-_61573003 | 0.02 |
ENSRNOT00000012179
|
Slc35b4
|
solute carrier family 35 member B4 |
| chr3_+_114087287 | 0.02 |
ENSRNOT00000023017
|
B2m
|
beta-2 microglobulin |
| chr10_-_18574909 | 0.02 |
ENSRNOT00000083090
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr8_+_60117729 | 0.02 |
ENSRNOT00000021074
|
Isl2
|
ISL LIM homeobox 2 |
| chr5_+_164950917 | 0.02 |
ENSRNOT00000083678
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr11_-_81757813 | 0.01 |
ENSRNOT00000002462
|
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr9_-_84894599 | 0.01 |
ENSRNOT00000018423
|
Hdac1l
|
histone deacetylase 1-like |
| chr20_+_3364814 | 0.01 |
ENSRNOT00000001077
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr16_-_81706664 | 0.01 |
ENSRNOT00000026580
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr8_-_62948720 | 0.01 |
ENSRNOT00000075476
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr9_+_82596355 | 0.01 |
ENSRNOT00000065076
|
Speg
|
SPEG complex locus |
| chr4_+_78458625 | 0.01 |
ENSRNOT00000049891
|
Tmem176a
|
transmembrane protein 176A |
| chr4_+_7355574 | 0.01 |
ENSRNOT00000013800
|
Kcnh2
|
potassium voltage-gated channel subfamily H member 2 |
| chr1_-_220096319 | 0.01 |
ENSRNOT00000091787
ENSRNOT00000073983 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr19_+_10142496 | 0.01 |
ENSRNOT00000088645
ENSRNOT00000060351 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr15_-_19876389 | 0.01 |
ENSRNOT00000012400
ENSRNOT00000086368 |
Fermt2
|
fermitin family member 2 |
| chr5_-_147743723 | 0.01 |
ENSRNOT00000012854
|
Hdac1
|
histone deacetylase 1 |
| chr5_+_164951789 | 0.01 |
ENSRNOT00000055658
ENSRNOT00000012193 |
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr19_-_55300403 | 0.01 |
ENSRNOT00000018591
|
Rnf166
|
ring finger protein 166 |
| chr1_-_219853329 | 0.01 |
ENSRNOT00000026169
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr5_-_74190991 | 0.01 |
ENSRNOT00000090366
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr1_+_29432152 | 0.01 |
ENSRNOT00000019436
ENSRNOT00000083276 |
Trmt11
|
tRNA methyltransferase 11 homolog |
| chr8_+_49342067 | 0.01 |
ENSRNOT00000021693
|
Mpzl2
|
myelin protein zero-like 2 |
| chr16_+_20109200 | 0.01 |
ENSRNOT00000079784
|
Jak3
|
Janus kinase 3 |
| chr20_+_4852496 | 0.01 |
ENSRNOT00000088936
|
Lta
|
lymphotoxin alpha |
| chr3_-_13978224 | 0.01 |
ENSRNOT00000025528
|
Phf19
|
PHD finger protein 19 |
| chr7_+_60099120 | 0.00 |
ENSRNOT00000007338
|
LOC100911101
|
leucine-rich repeat-containing protein 10-like |
| chr13_-_98478327 | 0.00 |
ENSRNOT00000030135
|
Coq8a
|
coenzyme Q8A |
| chr1_+_220446425 | 0.00 |
ENSRNOT00000027339
|
LOC100911932
|
endosialin-like |
| chr12_+_7454884 | 0.00 |
ENSRNOT00000077328
|
LOC100910196
|
katanin p60 ATPase-containing subunit A-like 1-like |
| chr16_-_21348391 | 0.00 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr6_-_47811853 | 0.00 |
ENSRNOT00000010942
|
Allc
|
allantoicase |
| chr3_+_151609602 | 0.00 |
ENSRNOT00000065052
|
Spag4
|
sperm associated antigen 4 |
| chr19_-_24831500 | 0.00 |
ENSRNOT00000005770
|
Pkn1
|
protein kinase N1 |
| chr2_+_27365148 | 0.00 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr15_-_46719252 | 0.00 |
ENSRNOT00000015452
|
Mtmr9
|
myotubularin related protein 9 |
| chr8_-_114892910 | 0.00 |
ENSRNOT00000074934
|
Ppm1m
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mtf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) astrocyte activation(GO:0048143) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.0 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.0 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.0 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |