Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
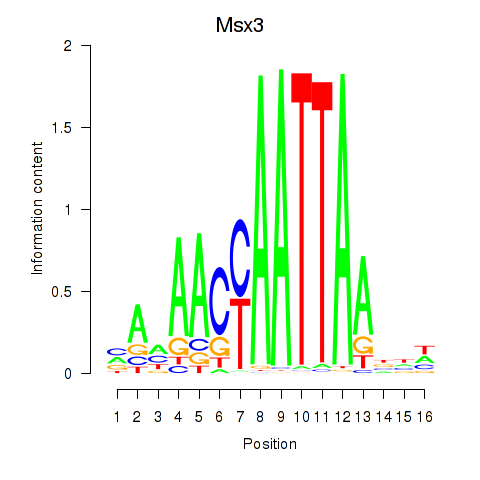
Results for Msx3
Z-value: 0.42
Transcription factors associated with Msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx3
|
ENSRNOG00000046776 | msh homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx3 | rn6_v1_chr1_-_212517151_212517151 | 0.13 | 8.3e-01 | Click! |
Activity profile of Msx3 motif
Sorted Z-values of Msx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrM_+_9870 | 0.13 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr3_-_90751055 | 0.12 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr4_+_147832136 | 0.12 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr5_+_6373583 | 0.12 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr6_-_76552559 | 0.12 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr9_+_73378057 | 0.11 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr2_-_219262901 | 0.11 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr3_-_165700489 | 0.10 |
ENSRNOT00000017008
|
Zfp93
|
zinc finger protein 93 |
| chr13_+_96303703 | 0.09 |
ENSRNOT00000084718
ENSRNOT00000029723 |
Efcab2
|
EF-hand calcium binding domain 2 |
| chr15_-_55209342 | 0.08 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr4_-_66955732 | 0.08 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chrX_+_14019961 | 0.07 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr12_-_45801842 | 0.07 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr11_-_71136673 | 0.06 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr4_-_176606382 | 0.06 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr8_+_48718329 | 0.06 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr13_-_76049363 | 0.06 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr3_-_10196626 | 0.06 |
ENSRNOT00000012234
|
Prdm12
|
PR/SET domain 12 |
| chr4_+_88694583 | 0.06 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr7_+_24638208 | 0.05 |
ENSRNOT00000009135
|
Mterf2
|
mitochondrial transcription termination factor 2 |
| chr15_+_87722221 | 0.05 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr2_-_196177775 | 0.05 |
ENSRNOT00000028553
|
Zfp687
|
zinc finger protein 687 |
| chr2_+_57276919 | 0.05 |
ENSRNOT00000063899
|
RGD1310081
|
similar to hypothetical protein FLJ13231 |
| chr2_-_178521038 | 0.05 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chrM_+_9451 | 0.05 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr6_-_42002819 | 0.05 |
ENSRNOT00000032417
|
Greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr2_+_195996521 | 0.05 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr18_+_45023932 | 0.04 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr16_+_84465656 | 0.04 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr1_-_241046249 | 0.04 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr7_-_120770435 | 0.04 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr4_+_31229913 | 0.04 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr7_+_143122269 | 0.04 |
ENSRNOT00000082542
ENSRNOT00000045495 ENSRNOT00000081386 ENSRNOT00000067422 |
Krt86
|
keratin 86 |
| chr18_-_4294136 | 0.03 |
ENSRNOT00000091909
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr12_-_22138382 | 0.03 |
ENSRNOT00000001899
|
Lrch4
|
leucine rich repeats and calponin homology domain containing 4 |
| chr4_+_176606649 | 0.03 |
ENSRNOT00000084700
|
Golt1b
|
golgi transport 1B |
| chr1_-_64147251 | 0.03 |
ENSRNOT00000088502
|
Cnot3
|
CCR4-NOT transcription complex, subunit 3 |
| chr8_-_39551700 | 0.03 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr17_-_87826421 | 0.03 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr1_+_217459215 | 0.03 |
ENSRNOT00000092386
ENSRNOT00000092357 |
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_+_260093641 | 0.03 |
ENSRNOT00000019521
|
Ccnj
|
cyclin J |
| chr13_+_98311827 | 0.03 |
ENSRNOT00000082844
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr1_+_101603222 | 0.03 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr2_+_220432037 | 0.03 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr7_-_72328128 | 0.03 |
ENSRNOT00000008227
|
Tspyl5
|
TSPY-like 5 |
| chr3_+_112346627 | 0.02 |
ENSRNOT00000074392
|
Snap23
|
synaptosomal-associated protein 23 |
| chr8_-_6305033 | 0.02 |
ENSRNOT00000029887
|
Cep126
|
centrosomal protein 126 |
| chr13_-_111972603 | 0.02 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chrX_+_118197217 | 0.02 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chrX_+_71342775 | 0.02 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr1_-_38538987 | 0.02 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr18_-_43945273 | 0.02 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr8_+_115546893 | 0.02 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr10_-_86645529 | 0.01 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr2_-_256154584 | 0.01 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr1_-_164101578 | 0.01 |
ENSRNOT00000022176
|
Uvrag
|
UV radiation resistance associated |
| chr15_+_39779648 | 0.01 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr5_+_133819726 | 0.01 |
ENSRNOT00000081075
|
Stil
|
Scl/Tal1 interrupting locus |
| chr7_-_143228060 | 0.01 |
ENSRNOT00000088923
ENSRNOT00000012640 |
Krt75
|
keratin 75 |
| chr15_-_36798814 | 0.01 |
ENSRNOT00000065764
|
Cenpj
|
centromere protein J |
| chr6_-_50618694 | 0.01 |
ENSRNOT00000008980
|
Dld
|
dihydrolipoamide dehydrogenase |
| chr3_-_165537940 | 0.01 |
ENSRNOT00000071119
ENSRNOT00000070964 |
Sall4
|
spalt-like transcription factor 4 |
| chr8_+_104106740 | 0.01 |
ENSRNOT00000015015
|
Tfdp2
|
transcription factor Dp-2 |
| chr5_-_56567320 | 0.00 |
ENSRNOT00000066081
|
Topors
|
TOP1 binding arginine/serine rich protein |
| chr7_+_42304534 | 0.00 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr8_+_79638696 | 0.00 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr6_-_111417813 | 0.00 |
ENSRNOT00000016324
ENSRNOT00000085458 |
Sptlc2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr2_-_9472384 | 0.00 |
ENSRNOT00000084295
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903944 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |