Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
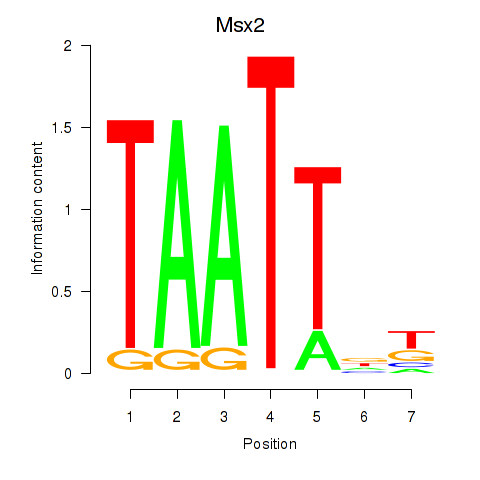
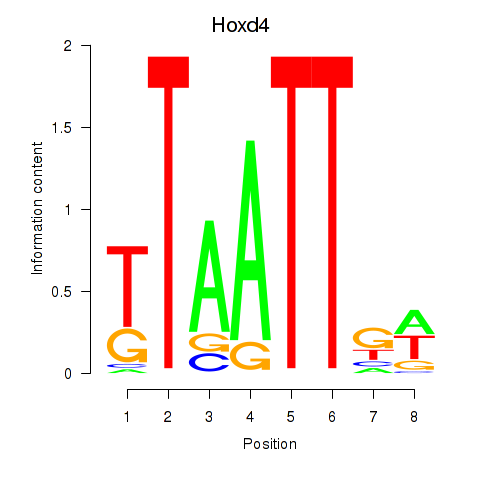
Results for Msx2_Hoxd4
Z-value: 0.27
Transcription factors associated with Msx2_Hoxd4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Msx2
|
ENSRNOG00000018355 | msh homeobox 2 |
|
Hoxd4
|
ENSRNOG00000001578 | homeo box D4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx2 | rn6_v1_chr17_+_11683862_11683862 | -0.41 | 4.9e-01 | Click! |
Activity profile of Msx2_Hoxd4 motif
Sorted Z-values of Msx2_Hoxd4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_86671515 | 0.07 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr2_+_72006099 | 0.06 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr6_-_76552559 | 0.06 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr9_-_30844199 | 0.06 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr2_-_31753528 | 0.05 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr20_+_25990656 | 0.05 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr19_+_25043680 | 0.05 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr5_-_168734296 | 0.05 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chrX_+_908044 | 0.04 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr15_+_56666012 | 0.04 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr1_-_166912524 | 0.04 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr13_-_76049363 | 0.04 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr6_+_64789940 | 0.03 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr6_+_27887797 | 0.03 |
ENSRNOT00000015827
|
Asxl2
|
additional sex combs like 2, transcriptional regulator |
| chr14_+_38192870 | 0.03 |
ENSRNOT00000077080
|
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr9_+_52687868 | 0.03 |
ENSRNOT00000083864
|
Wdr75
|
WD repeat domain 75 |
| chr19_-_22194740 | 0.03 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_+_47032113 | 0.03 |
ENSRNOT00000024412
|
Tulp4
|
tubby like protein 4 |
| chr11_-_29710849 | 0.03 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr3_-_25212049 | 0.03 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr2_-_35104963 | 0.03 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr18_-_1828606 | 0.03 |
ENSRNOT00000033312
|
Esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr2_+_127489771 | 0.03 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr9_-_63641400 | 0.03 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr15_-_71779033 | 0.03 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chrX_+_84064427 | 0.03 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr3_-_90751055 | 0.02 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr2_-_33025271 | 0.02 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr13_+_46169963 | 0.02 |
ENSRNOT00000005212
|
Thsd7b
|
thrombospondin type 1 domain containing 7B |
| chr11_-_90406797 | 0.02 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr13_+_90533365 | 0.02 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr8_+_70317586 | 0.02 |
ENSRNOT00000016260
|
Dennd4a
|
DENN domain containing 4A |
| chr7_+_122818975 | 0.02 |
ENSRNOT00000000206
|
Ep300
|
E1A binding protein p300 |
| chr12_-_35979193 | 0.02 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr2_-_30634243 | 0.02 |
ENSRNOT00000077537
|
Marveld2
|
MARVEL domain containing 2 |
| chr5_+_104698040 | 0.02 |
ENSRNOT00000009127
|
Adamtsl1
|
ADAMTS-like 1 |
| chr20_+_27975549 | 0.02 |
ENSRNOT00000092075
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr14_+_48768537 | 0.02 |
ENSRNOT00000082599
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr7_+_42304534 | 0.02 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr2_+_196013799 | 0.02 |
ENSRNOT00000084023
|
Pogz
|
pogo transposable element with ZNF domain |
| chr5_-_154363329 | 0.02 |
ENSRNOT00000064596
|
Eloa
|
elongin A |
| chr16_-_54137660 | 0.02 |
ENSRNOT00000085435
|
Pcm1
|
pericentriolar material 1 |
| chr19_+_32308236 | 0.02 |
ENSRNOT00000015508
|
Mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr2_-_196377883 | 0.02 |
ENSRNOT00000028661
|
Gabpb2
|
GA binding protein transcription factor, beta subunit 2 |
| chr19_+_43163129 | 0.01 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr8_-_96266342 | 0.01 |
ENSRNOT00000078891
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr6_-_94834908 | 0.01 |
ENSRNOT00000006284
|
L3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr6_+_69971227 | 0.01 |
ENSRNOT00000075349
|
Foxg1
|
forkhead box G1 |
| chr10_-_90940118 | 0.01 |
ENSRNOT00000093143
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr11_-_782954 | 0.01 |
ENSRNOT00000040065
|
Epha3
|
Eph receptor A3 |
| chr3_-_52447622 | 0.01 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr5_-_48436531 | 0.01 |
ENSRNOT00000060914
|
Pm20d2
|
peptidase M20 domain containing 2 |
| chr17_-_53713408 | 0.01 |
ENSRNOT00000022445
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chrX_-_106607352 | 0.01 |
ENSRNOT00000082858
|
AABR07040624.1
|
|
| chrM_+_8599 | 0.01 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr14_-_45859908 | 0.01 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr2_-_181531978 | 0.01 |
ENSRNOT00000072029
|
Npy2r
|
neuropeptide Y receptor Y2 |
| chr4_-_129619142 | 0.01 |
ENSRNOT00000047453
|
Lmod3
|
leiomodin 3 |
| chr11_+_61605937 | 0.01 |
ENSRNOT00000093455
ENSRNOT00000093242 |
Gramd1c
|
GRAM domain containing 1C |
| chr2_+_165805144 | 0.01 |
ENSRNOT00000073716
|
Rabif
|
RAB interacting factor |
| chr2_-_210766501 | 0.01 |
ENSRNOT00000025800
|
Gstm3
|
glutathione S-transferase mu 3 |
| chr20_+_9948908 | 0.01 |
ENSRNOT00000001541
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chr2_-_219262901 | 0.00 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr10_-_87521514 | 0.00 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr5_-_12172009 | 0.00 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr16_+_50049828 | 0.00 |
ENSRNOT00000034448
|
Fam149a
|
family with sequence similarity 149, member A |
| chr2_+_54466280 | 0.00 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chrM_+_7006 | 0.00 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr10_+_65733991 | 0.00 |
ENSRNOT00000013698
|
Slc46a1
|
solute carrier family 46 member 1 |
| chrM_+_7919 | 0.00 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr1_+_22364551 | 0.00 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr16_+_61926586 | 0.00 |
ENSRNOT00000058884
|
AABR07026059.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Msx2_Hoxd4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |