Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Mnt
Z-value: 0.76
Transcription factors associated with Mnt
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mnt
|
ENSRNOG00000002894 | MAX network transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnt | rn6_v1_chr10_+_61685241_61685241 | -0.43 | 4.8e-01 | Click! |
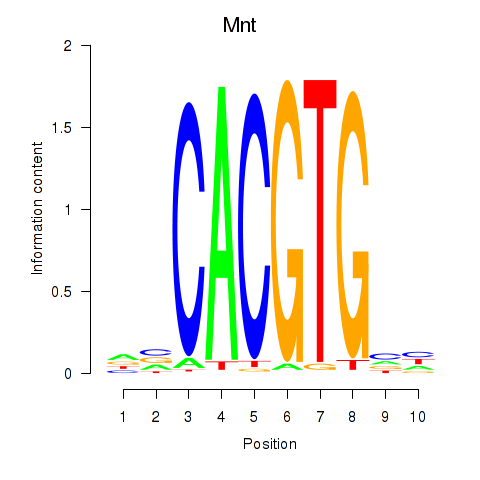
Activity profile of Mnt motif
Sorted Z-values of Mnt motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_123119460 | 1.05 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chr2_-_197991198 | 0.73 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_197991574 | 0.70 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr17_-_9695292 | 0.63 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr2_-_187160373 | 0.53 |
ENSRNOT00000018961
|
Ntrk1
|
neurotrophic receptor tyrosine kinase 1 |
| chr7_-_13751271 | 0.45 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chr14_+_4362717 | 0.44 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr1_+_33910912 | 0.35 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr11_-_36479868 | 0.35 |
ENSRNOT00000075762
|
LOC100911295
|
non-histone chromosomal protein HMG-14-like |
| chr16_+_49462889 | 0.32 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr3_-_66417741 | 0.31 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_-_44746549 | 0.30 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr1_+_163445527 | 0.28 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr13_-_90814119 | 0.28 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr15_+_37790141 | 0.26 |
ENSRNOT00000076392
ENSRNOT00000091953 |
Il17d
|
interleukin 17D |
| chr3_-_8852192 | 0.25 |
ENSRNOT00000022786
|
Dolk
|
dolichol kinase |
| chr12_-_16934706 | 0.25 |
ENSRNOT00000001720
|
Mafk
|
MAF bZIP transcription factor K |
| chr2_+_127538659 | 0.25 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr19_+_14508616 | 0.24 |
ENSRNOT00000019192
|
Hmox1
|
heme oxygenase 1 |
| chr4_-_123118186 | 0.24 |
ENSRNOT00000038096
|
LOC100361898
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr9_-_16848503 | 0.24 |
ENSRNOT00000024815
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr5_-_147953093 | 0.24 |
ENSRNOT00000075270
|
Khdrbs1
|
KH RNA binding domain containing, signal transduction associated 1 |
| chr20_-_5533448 | 0.23 |
ENSRNOT00000000568
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr1_+_73719005 | 0.23 |
ENSRNOT00000025110
|
Cdc42ep5
|
CDC42 effector protein 5 |
| chr3_+_113415774 | 0.23 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr4_+_33638709 | 0.22 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr20_-_5533600 | 0.22 |
ENSRNOT00000072319
|
Cuta
|
cutA divalent cation tolerance homolog |
| chr20_+_13732198 | 0.22 |
ENSRNOT00000008608
|
LOC103694877
|
macrophage migration inhibitory factor |
| chr6_+_33885495 | 0.21 |
ENSRNOT00000086633
|
Sdc1
|
syndecan 1 |
| chr17_+_16333415 | 0.21 |
ENSRNOT00000060550
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr4_-_82173207 | 0.21 |
ENSRNOT00000074167
|
Hoxa5
|
homeo box A5 |
| chr9_-_97065817 | 0.20 |
ENSRNOT00000026419
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr3_-_120306551 | 0.20 |
ENSRNOT00000021082
|
Mall
|
mal, T-cell differentiation protein-like |
| chr19_+_55669626 | 0.20 |
ENSRNOT00000033352
|
Cdh15
|
cadherin 15 |
| chr8_-_66863476 | 0.20 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chr5_+_150725654 | 0.20 |
ENSRNOT00000089852
ENSRNOT00000017740 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr2_+_189400696 | 0.20 |
ENSRNOT00000046919
ENSRNOT00000089801 |
LOC361985
|
similar to NICE-3 |
| chr20_-_13706205 | 0.19 |
ENSRNOT00000038623
|
Derl3
|
derlin 3 |
| chr3_+_147643250 | 0.19 |
ENSRNOT00000000013
|
Tcf15
|
transcription factor 15 |
| chr10_+_84152152 | 0.19 |
ENSRNOT00000010724
|
Hoxb5
|
homeo box B5 |
| chr1_+_261229347 | 0.19 |
ENSRNOT00000018485
|
Ubtd1
|
ubiquitin domain containing 1 |
| chr1_-_91042230 | 0.19 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr8_+_117117430 | 0.19 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr10_+_91710495 | 0.18 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr6_+_42852683 | 0.18 |
ENSRNOT00000079185
|
Odc1
|
ornithine decarboxylase 1 |
| chr1_-_24191908 | 0.18 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_-_18683440 | 0.18 |
ENSRNOT00000068323
|
Rps28
|
ribosomal protein S28 |
| chr1_-_255376833 | 0.18 |
ENSRNOT00000024941
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr10_-_13536899 | 0.18 |
ENSRNOT00000008537
|
Amdhd2
|
amidohydrolase domain containing 2 |
| chrX_-_32355296 | 0.17 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr8_-_104221342 | 0.17 |
ENSRNOT00000015476
|
Atp1b3
|
ATPase Na+/K+ transporting subunit beta 3 |
| chr9_+_98490608 | 0.17 |
ENSRNOT00000027232
|
Klhl30
|
kelch-like family member 30 |
| chr4_-_165629996 | 0.17 |
ENSRNOT00000068185
ENSRNOT00000007427 |
Ybx3
|
Y box binding protein 3 |
| chr5_+_150080072 | 0.16 |
ENSRNOT00000047917
|
Tmem200b
|
transmembrane protein 200B |
| chr5_-_152464850 | 0.16 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chr1_+_88686731 | 0.16 |
ENSRNOT00000028222
|
Polr2i
|
RNA polymerase II subunit I |
| chr10_+_55940533 | 0.15 |
ENSRNOT00000012061
|
RGD1563441
|
similar to RIKEN cDNA A030009H04 |
| chr16_+_80826681 | 0.15 |
ENSRNOT00000000123
|
Coprs
|
coordinator of PRMT5 and differentiation stimulator |
| chr10_+_16542882 | 0.15 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr10_+_84135116 | 0.15 |
ENSRNOT00000031035
|
Hoxb7
|
homeo box B7 |
| chr7_+_3332788 | 0.15 |
ENSRNOT00000010180
|
Cd63
|
Cd63 molecule |
| chr9_+_10446699 | 0.15 |
ENSRNOT00000075344
|
LOC301124
|
hypothetical LOC301124 |
| chr3_+_61685619 | 0.14 |
ENSRNOT00000002140
|
Hoxd1
|
homeo box D1 |
| chr3_-_81282157 | 0.14 |
ENSRNOT00000051258
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr11_-_60613718 | 0.14 |
ENSRNOT00000002906
|
Atg3
|
autophagy related 3 |
| chr2_+_189106039 | 0.14 |
ENSRNOT00000028210
|
Ube2q1
|
ubiquitin conjugating enzyme E2 Q1 |
| chr8_-_61917125 | 0.14 |
ENSRNOT00000085049
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr7_-_140770647 | 0.14 |
ENSRNOT00000081784
|
C1ql4
|
complement C1q like 4 |
| chr10_-_81587858 | 0.14 |
ENSRNOT00000003582
|
Utp18
|
UTP18 small subunit processome component |
| chr1_-_78333321 | 0.14 |
ENSRNOT00000020402
|
Sae1
|
SUMO1 activating enzyme subunit 1 |
| chr16_-_81706664 | 0.14 |
ENSRNOT00000026580
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr7_+_18683553 | 0.14 |
ENSRNOT00000009425
|
Ndufa7
|
NADH:ubiquinone oxidoreductase subunit A7 |
| chr19_+_52225582 | 0.13 |
ENSRNOT00000020917
|
Dnaaf1
|
dynein, axonemal, assembly factor 1 |
| chr10_+_16970626 | 0.13 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr5_-_136721379 | 0.13 |
ENSRNOT00000026704
|
Atp6v0b
|
ATPase H+ transporting V0 subunit B |
| chr1_-_226255886 | 0.13 |
ENSRNOT00000027842
|
Fen1
|
flap structure-specific endonuclease 1 |
| chr7_+_128500011 | 0.13 |
ENSRNOT00000074625
|
Fam19a5
|
family with sequence similarity 19 member A5, C-C motif chemokine like |
| chr1_+_240776969 | 0.13 |
ENSRNOT00000049732
|
RGD1561453
|
similar to ribosomal protein S12 |
| chr11_+_39482408 | 0.13 |
ENSRNOT00000075126
|
Hmgn1
|
high mobility group nucleosome binding domain 1 |
| chr10_-_57618527 | 0.13 |
ENSRNOT00000037517
|
C1qbp
|
complement C1q binding protein |
| chr1_+_12823363 | 0.13 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr11_+_47090245 | 0.13 |
ENSRNOT00000060645
|
AABR07033987.1
|
|
| chr17_+_54280851 | 0.13 |
ENSRNOT00000024022
|
Arhgap12
|
Rho GTPase activating protein 12 |
| chr1_-_214455039 | 0.13 |
ENSRNOT00000071028
|
Polr2l
|
RNA polymerase II subunit L |
| chr1_-_88686615 | 0.12 |
ENSRNOT00000070921
|
Tbcb
|
tubulin folding cofactor B |
| chr3_+_147358858 | 0.12 |
ENSRNOT00000012951
|
Rspo4
|
R-spondin 4 |
| chr5_-_166726794 | 0.12 |
ENSRNOT00000022799
|
Slc25a33
|
solute carrier family 25 member 33 |
| chr10_+_83476107 | 0.12 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr9_-_113331319 | 0.12 |
ENSRNOT00000020681
|
Vapa
|
VAMP associated protein A |
| chr1_+_40816107 | 0.11 |
ENSRNOT00000060767
|
Akap12
|
A-kinase anchoring protein 12 |
| chr1_-_101457126 | 0.11 |
ENSRNOT00000087061
ENSRNOT00000028328 |
Bax
|
BCL2 associated X, apoptosis regulator |
| chr1_-_101123402 | 0.11 |
ENSRNOT00000027976
|
Rpl13a
|
|
| chr16_+_69048730 | 0.11 |
ENSRNOT00000086082
ENSRNOT00000078128 |
Rab11fip1
|
RAB11 family interacting protein 1 |
| chr8_+_39305128 | 0.11 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr11_-_34598102 | 0.11 |
ENSRNOT00000068743
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr19_+_58565576 | 0.11 |
ENSRNOT00000026965
|
Ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr1_+_82452469 | 0.11 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr14_-_11198194 | 0.11 |
ENSRNOT00000003083
|
Enoph1
|
enolase-phosphatase 1 |
| chr17_+_9837402 | 0.11 |
ENSRNOT00000076436
|
Rab24
|
RAB24, member RAS oncogene family |
| chr1_-_84008293 | 0.11 |
ENSRNOT00000002053
|
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr12_+_19573909 | 0.11 |
ENSRNOT00000029279
|
Lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr7_+_121480723 | 0.11 |
ENSRNOT00000065304
|
Atf4
|
activating transcription factor 4 |
| chr9_+_82345719 | 0.11 |
ENSRNOT00000057330
ENSRNOT00000078260 |
Fam134a
|
family with sequence similarity 134, member A |
| chr3_-_173953684 | 0.11 |
ENSRNOT00000090468
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr1_-_64021321 | 0.10 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr16_-_75855745 | 0.10 |
ENSRNOT00000031291
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 |
| chr10_-_13187461 | 0.10 |
ENSRNOT00000007039
|
Prss41
|
protease, serine, 41 |
| chr11_-_83985385 | 0.10 |
ENSRNOT00000002334
|
LOC102551435
|
endothelin-converting enzyme 2-like |
| chr7_+_143882000 | 0.10 |
ENSRNOT00000017110
ENSRNOT00000091053 |
Mfsd5
|
major facilitator superfamily domain containing 5 |
| chr5_+_171242897 | 0.10 |
ENSRNOT00000032859
|
RGD1304567
|
similar to RIKEN cDNA A430005L14 |
| chr6_+_136150785 | 0.10 |
ENSRNOT00000078211
ENSRNOT00000015308 |
LOC103692719
|
tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A |
| chr7_+_11521998 | 0.10 |
ENSRNOT00000026960
|
Sgta
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr5_-_157820889 | 0.10 |
ENSRNOT00000024326
|
Mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr7_+_77966722 | 0.10 |
ENSRNOT00000006142
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr2_+_188489362 | 0.10 |
ENSRNOT00000027815
|
Clk2
|
CDC-like kinase 2 |
| chr10_-_89959176 | 0.10 |
ENSRNOT00000028264
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr5_-_138470096 | 0.10 |
ENSRNOT00000011392
|
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr20_-_5723902 | 0.10 |
ENSRNOT00000036871
|
Uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr7_+_121889157 | 0.10 |
ENSRNOT00000024835
|
Fam83f
|
family with sequence similarity 83, member F |
| chr3_-_161812243 | 0.10 |
ENSRNOT00000025238
ENSRNOT00000087646 |
Slc35c2
|
solute carrier family 35 member C2 |
| chr1_-_224986291 | 0.10 |
ENSRNOT00000026284
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr1_-_198706852 | 0.09 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr17_-_9837293 | 0.09 |
ENSRNOT00000091320
ENSRNOT00000022134 |
Prelid1
|
PRELI domain containing 1 |
| chr14_-_84189266 | 0.09 |
ENSRNOT00000005934
|
Tcn2
|
transcobalamin 2 |
| chr10_+_103438303 | 0.09 |
ENSRNOT00000085262
|
Cd300a
|
Cd300a molecule |
| chr10_-_86509454 | 0.09 |
ENSRNOT00000009454
|
Ikzf3
|
IKAROS family zinc finger 3 |
| chr5_-_14356692 | 0.09 |
ENSRNOT00000085654
|
Atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr7_-_117788550 | 0.09 |
ENSRNOT00000021775
|
MGC94207
|
similar to RIKEN cDNA C030006K11 |
| chr7_-_3346238 | 0.09 |
ENSRNOT00000010380
|
Bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr12_+_12738812 | 0.09 |
ENSRNOT00000092233
ENSRNOT00000001379 |
Aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr4_+_78981987 | 0.09 |
ENSRNOT00000012490
|
Ccdc126
|
coiled-coil domain containing 126 |
| chr4_-_157679962 | 0.09 |
ENSRNOT00000050443
|
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr18_-_81539065 | 0.08 |
ENSRNOT00000021400
|
Cndp2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr3_+_72226613 | 0.08 |
ENSRNOT00000010619
|
Timm10
|
translocase of inner mitochondrial membrane 10 |
| chr9_-_9702306 | 0.08 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chrX_+_62363953 | 0.08 |
ENSRNOT00000083362
|
Arx
|
aristaless related homeobox |
| chr10_-_40375605 | 0.08 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chr11_-_34598275 | 0.08 |
ENSRNOT00000077233
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr7_-_73216251 | 0.08 |
ENSRNOT00000057587
|
LOC100362027
|
ribosomal protein L30-like |
| chr1_-_199159125 | 0.08 |
ENSRNOT00000025618
|
Bcl7c
|
BCL tumor suppressor 7C |
| chr9_+_69953440 | 0.08 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_100393303 | 0.08 |
ENSRNOT00000026251
|
Syt3
|
synaptotagmin 3 |
| chr5_-_168004724 | 0.08 |
ENSRNOT00000024711
|
Park7
|
Parkinsonism associated deglycase |
| chr10_-_81653717 | 0.08 |
ENSRNOT00000003611
|
Nme2
|
NME/NM23 nucleoside diphosphate kinase 2 |
| chr1_-_82452281 | 0.08 |
ENSRNOT00000027995
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr10_-_89958805 | 0.08 |
ENSRNOT00000084266
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr20_-_6961162 | 0.08 |
ENSRNOT00000000635
|
Mtch1
|
mitochondrial carrier 1 |
| chr7_-_73270308 | 0.08 |
ENSRNOT00000007430
|
Rida
|
reactive intermediate imine deaminase A homolog |
| chrX_+_43497763 | 0.08 |
ENSRNOT00000005014
|
Prdx4
|
peroxiredoxin 4 |
| chr4_+_6827429 | 0.08 |
ENSRNOT00000071737
|
Rheb
|
Ras homolog enriched in brain |
| chr11_+_80255790 | 0.08 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr8_+_50559126 | 0.08 |
ENSRNOT00000024918
|
Apoa5
|
apolipoprotein A5 |
| chr1_+_221773254 | 0.08 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr9_-_16612136 | 0.08 |
ENSRNOT00000023495
|
Mea1
|
male-enhanced antigen 1 |
| chr7_-_18554227 | 0.08 |
ENSRNOT00000036172
ENSRNOT00000011047 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr12_+_22229079 | 0.08 |
ENSRNOT00000001911
|
Gnb2
|
G protein subunit beta 2 |
| chr10_-_83898527 | 0.08 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr4_-_100099517 | 0.08 |
ENSRNOT00000014277
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr3_+_11424099 | 0.08 |
ENSRNOT00000019184
|
Ptges2
|
prostaglandin E synthase 2 |
| chr5_+_169506138 | 0.08 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr4_-_82258765 | 0.08 |
ENSRNOT00000008523
|
Hoxa5
|
homeo box A5 |
| chr10_+_82375572 | 0.08 |
ENSRNOT00000004947
|
Mrpl27
|
mitochondrial ribosomal protein L27 |
| chr1_+_222229835 | 0.08 |
ENSRNOT00000051749
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_+_170578889 | 0.08 |
ENSRNOT00000025906
|
Ilk
|
integrin-linked kinase |
| chr19_-_49532811 | 0.08 |
ENSRNOT00000015967
|
Gcsh
|
glycine cleavage system protein H |
| chr10_+_103266296 | 0.07 |
ENSRNOT00000038852
|
Dnai2
|
dynein, axonemal, intermediate chain 2 |
| chr18_+_70996044 | 0.07 |
ENSRNOT00000024950
ENSRNOT00000066336 |
Dym
|
dymeclin |
| chrX_-_156399760 | 0.07 |
ENSRNOT00000086921
|
Fam50a
|
family with sequence similarity 50, member A |
| chr1_+_173532803 | 0.07 |
ENSRNOT00000021017
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr17_+_85356042 | 0.07 |
ENSRNOT00000022201
|
Commd3
|
COMM domain containing 3 |
| chr14_-_33580566 | 0.07 |
ENSRNOT00000063942
|
Paics
|
phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase |
| chr13_+_90514336 | 0.07 |
ENSRNOT00000088996
ENSRNOT00000085377 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr5_-_171312026 | 0.07 |
ENSRNOT00000063779
|
Smim1
|
small integral membrane protein 1 |
| chr5_-_135628750 | 0.07 |
ENSRNOT00000079236
|
AC126292.3
|
|
| chr1_-_265899958 | 0.07 |
ENSRNOT00000026013
|
Pitx3
|
paired-like homeodomain 3 |
| chr10_-_56531483 | 0.07 |
ENSRNOT00000022343
|
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr2_+_190007216 | 0.07 |
ENSRNOT00000015612
|
S100a6
|
S100 calcium binding protein A6 |
| chr9_-_66877972 | 0.07 |
ENSRNOT00000023463
|
Wdr12
|
WD repeat domain 12 |
| chr1_-_164143818 | 0.07 |
ENSRNOT00000022557
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr4_+_342302 | 0.07 |
ENSRNOT00000009233
|
Insig1
|
insulin induced gene 1 |
| chr1_-_93949187 | 0.07 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr1_+_216293087 | 0.07 |
ENSRNOT00000027875
ENSRNOT00000087153 |
Kcnq1
|
potassium voltage-gated channel subfamily Q member 1 |
| chr4_+_7257752 | 0.07 |
ENSRNOT00000088357
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr16_-_60427474 | 0.07 |
ENSRNOT00000051720
|
Ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr3_-_12944494 | 0.07 |
ENSRNOT00000023172
|
Mvb12b
|
multivesicular body subunit 12B |
| chr1_-_100980340 | 0.07 |
ENSRNOT00000064272
|
Prmt1
|
protein arginine methyltransferase 1 |
| chr15_-_45376476 | 0.07 |
ENSRNOT00000065838
|
Dleu7
|
deleted in lymphocytic leukemia, 7 |
| chr12_-_9864791 | 0.07 |
ENSRNOT00000073026
|
Gtf3a
|
general transcription factor III A |
| chr2_+_115337439 | 0.07 |
ENSRNOT00000015779
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr4_+_99937558 | 0.06 |
ENSRNOT00000050249
|
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr1_+_77994203 | 0.06 |
ENSRNOT00000002044
|
Napa
|
NSF attachment protein alpha |
| chr5_+_152681101 | 0.06 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr2_+_243105674 | 0.06 |
ENSRNOT00000013919
|
H2afz
|
H2A histone family, member Z |
| chr12_+_22451596 | 0.06 |
ENSRNOT00000072838
|
Trip6
|
thyroid hormone receptor interactor 6 |
| chr7_+_71294140 | 0.06 |
ENSRNOT00000087143
|
Ptdss1
|
phosphatidylserine synthase 1 |
| chr20_+_31265483 | 0.06 |
ENSRNOT00000079584
ENSRNOT00000000674 |
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr9_+_17817721 | 0.06 |
ENSRNOT00000086986
ENSRNOT00000026920 |
Hsp90ab1
|
heat shock protein 90 alpha family class B member 1 |
| chr12_+_23544287 | 0.06 |
ENSRNOT00000001938
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr1_-_163328591 | 0.06 |
ENSRNOT00000034843
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr4_-_82160240 | 0.06 |
ENSRNOT00000038775
ENSRNOT00000058985 |
Hoxa4
|
homeo box A4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mnt
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.5 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.3 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.1 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:0072268 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.3 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.1 | 0.2 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.3 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.1 | 1.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.2 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.0 | 0.1 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:1990737 | negative regulation of translational initiation in response to stress(GO:0032057) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0060101 | negative regulation of phagocytosis, engulfment(GO:0060101) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:1903195 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:1990790 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.0 | 0.2 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.1 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0045901 | regulation of translational termination(GO:0006449) positive regulation of translational elongation(GO:0045901) |
| 0.0 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.0 | GO:0080033 | cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:1904585 | response to sodium phosphate(GO:1904383) response to putrescine(GO:1904585) cellular response to putrescine(GO:1904586) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0018201 | N-terminal protein amino acid methylation(GO:0006480) peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.0 | GO:0051695 | actin filament uncapping(GO:0051695) positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.0 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.0 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.0 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 0.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.0 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.0 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.1 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:1990826 | nucleoplasmic periphery of the nuclear pore complex(GO:1990826) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.1 | 0.2 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 1.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.0 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.2 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.0 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |