Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
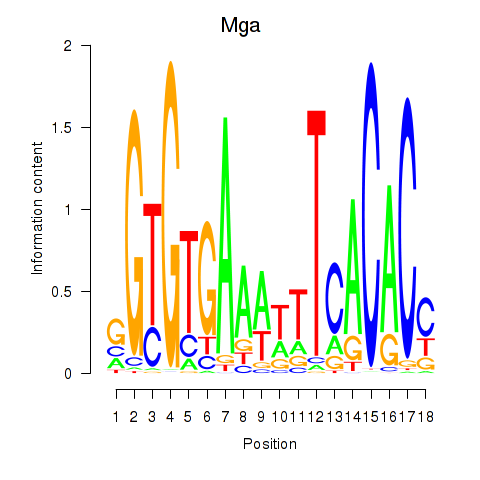
Results for Mga
Z-value: 0.28
Transcription factors associated with Mga
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mga
|
ENSRNOG00000006378 | MGA, MAX dimerization protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mga | rn6_v1_chr3_+_111699021_111699087 | 0.15 | 8.1e-01 | Click! |
Activity profile of Mga motif
Sorted Z-values of Mga motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_72540538 | 0.09 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr17_-_78910671 | 0.08 |
ENSRNOT00000064609
|
Acbd7
|
acyl-CoA binding domain containing 7 |
| chr7_+_119820537 | 0.06 |
ENSRNOT00000077256
ENSRNOT00000056221 |
Cyth4
|
cytohesin 4 |
| chr10_+_105498728 | 0.06 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr14_+_104250617 | 0.06 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr16_-_81434363 | 0.05 |
ENSRNOT00000080963
|
Rasa3
|
RAS p21 protein activator 3 |
| chr12_+_31530699 | 0.05 |
ENSRNOT00000033379
ENSRNOT00000067867 |
Rimbp2
|
RIMS binding protein 2 |
| chr1_+_282134981 | 0.05 |
ENSRNOT00000036203
|
Nanos1
|
nanos C2HC-type zinc finger 1 |
| chr7_-_117680004 | 0.04 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr4_+_163162211 | 0.04 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr17_+_43632397 | 0.04 |
ENSRNOT00000013790
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr18_-_70924708 | 0.04 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr2_-_195678848 | 0.04 |
ENSRNOT00000028303
ENSRNOT00000075569 |
Oaz3
|
ornithine decarboxylase antizyme 3 |
| chr1_+_29191192 | 0.03 |
ENSRNOT00000018718
|
Hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr1_+_65781547 | 0.03 |
ENSRNOT00000026204
|
Zfp329
|
zinc finger protein 329 |
| chr7_+_120176530 | 0.03 |
ENSRNOT00000087548
|
Triobp
|
TRIO and F-actin binding protein |
| chr6_+_106496992 | 0.03 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr7_+_58366192 | 0.03 |
ENSRNOT00000081891
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr7_+_25808419 | 0.03 |
ENSRNOT00000091571
|
Rfx4
|
regulatory factor X4 |
| chr6_-_123405246 | 0.03 |
ENSRNOT00000083665
|
Foxn3
|
forkhead box N3 |
| chr8_-_50526843 | 0.03 |
ENSRNOT00000092188
|
AABR07070085.1
|
|
| chr6_+_132242328 | 0.03 |
ENSRNOT00000081088
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr5_+_119727839 | 0.03 |
ENSRNOT00000014354
|
Cachd1
|
cache domain containing 1 |
| chr1_+_127604197 | 0.03 |
ENSRNOT00000018463
|
Lins1
|
lines homolog 1 |
| chr19_-_17494516 | 0.03 |
ENSRNOT00000078786
ENSRNOT00000076138 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chrX_+_45637415 | 0.03 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr2_+_187512164 | 0.03 |
ENSRNOT00000051394
|
Mef2d
|
myocyte enhancer factor 2D |
| chrX_-_1786978 | 0.03 |
ENSRNOT00000011317
|
Rbm10
|
RNA binding motif protein 10 |
| chr10_-_110232843 | 0.02 |
ENSRNOT00000054934
|
Cd7
|
Cd7 molecule |
| chr4_+_152437966 | 0.02 |
ENSRNOT00000013070
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr7_+_126096793 | 0.02 |
ENSRNOT00000047234
ENSRNOT00000088042 |
Fbln1
|
fibulin 1 |
| chr9_+_82632230 | 0.02 |
ENSRNOT00000085165
|
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr8_-_114449956 | 0.02 |
ENSRNOT00000056414
|
Col6a6
|
collagen type VI alpha 6 chain |
| chr10_+_105500290 | 0.02 |
ENSRNOT00000079080
ENSRNOT00000083593 |
Sphk1
|
sphingosine kinase 1 |
| chr2_+_188141350 | 0.02 |
ENSRNOT00000078913
|
Gon4l
|
gon-4 like |
| chr8_-_132027006 | 0.02 |
ENSRNOT00000050963
|
LOC367195
|
similar to 60S RIBOSOMAL PROTEIN L7 |
| chr1_-_101095594 | 0.02 |
ENSRNOT00000027944
|
Fcgrt
|
Fc fragment of IgG receptor and transporter |
| chr9_+_82632384 | 0.02 |
ENSRNOT00000027064
|
Gmppa
|
GDP-mannose pyrophosphorylase A |
| chr8_+_16287804 | 0.02 |
ENSRNOT00000024691
|
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr10_-_31359699 | 0.02 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr8_-_107656861 | 0.02 |
ENSRNOT00000050374
|
Mras
|
muscle RAS oncogene homolog |
| chr1_-_256013495 | 0.02 |
ENSRNOT00000023342
|
Ide
|
insulin degrading enzyme |
| chr4_+_181481147 | 0.01 |
ENSRNOT00000002523
|
Klhl42
|
kelch-like family, member 42 |
| chr20_-_5618254 | 0.01 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr9_-_65442257 | 0.01 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chr3_-_177095597 | 0.01 |
ENSRNOT00000029386
|
Samd10
|
sterile alpha motif domain containing 10 |
| chr6_+_113898420 | 0.01 |
ENSRNOT00000064872
|
Nrxn3
|
neurexin 3 |
| chr2_-_240611560 | 0.01 |
ENSRNOT00000074632
|
Cisd2
|
CDGSH iron sulfur domain 2 |
| chr4_+_85427555 | 0.01 |
ENSRNOT00000015469
|
Fam188b
|
family with sequence similarity 188, member B |
| chr14_+_75880410 | 0.01 |
ENSRNOT00000014078
|
Hs3st1
|
heparan sulfate-glucosamine 3-sulfotransferase 1 |
| chr5_+_69833225 | 0.01 |
ENSRNOT00000013759
|
Nipsnap3b
|
nipsnap homolog 3B |
| chr7_+_61165640 | 0.01 |
ENSRNOT00000087955
|
Mdm1
|
Mdm1 nuclear protein |
| chr20_-_14393793 | 0.01 |
ENSRNOT00000001764
|
Specc1l
|
sperm antigen with calponin homology and coiled-coil domains 1-like |
| chr4_-_78342863 | 0.01 |
ENSRNOT00000049038
|
Gimap6
|
GTPase, IMAP family member 6 |
| chr10_-_56167426 | 0.01 |
ENSRNOT00000013955
|
Efnb3
|
ephrin B3 |
| chr8_+_131845696 | 0.01 |
ENSRNOT00000005440
|
Tcaim
|
T cell activation inhibitor, mitochondrial |
| chr1_-_90151405 | 0.01 |
ENSRNOT00000028688
|
RGD1308428
|
similar to RIKEN cDNA 4931406P16 |
| chr1_-_47477524 | 0.01 |
ENSRNOT00000067916
|
Rsph3
|
radial spoke 3 homolog |
| chr20_-_7219548 | 0.01 |
ENSRNOT00000033907
|
Rps10
|
ribosomal protein S10 |
| chr2_+_155076464 | 0.01 |
ENSRNOT00000057619
|
LOC691033
|
similar to GTPase activating protein testicular GAP1 |
| chrX_-_118513061 | 0.01 |
ENSRNOT00000043338
|
Il13ra2
|
interleukin 13 receptor subunit alpha 2 |
| chr5_+_68717519 | 0.01 |
ENSRNOT00000066226
|
Smc2
|
structural maintenance of chromosomes 2 |
| chr12_-_2610917 | 0.01 |
ENSRNOT00000034124
|
Evi5l
|
ecotropic viral integration site 5-like |
| chr8_-_22929294 | 0.01 |
ENSRNOT00000015609
|
Rab3d
|
RAB3D, member RAS oncogene family |
| chr1_-_81152384 | 0.01 |
ENSRNOT00000026263
ENSRNOT00000077305 |
Zfp94
|
zinc finger protein 94 |
| chr16_+_80729400 | 0.01 |
ENSRNOT00000036383
|
Tdrp
|
testis development related protein |
| chr8_+_5768811 | 0.01 |
ENSRNOT00000013936
|
Mmp8
|
matrix metallopeptidase 8 |
| chr19_-_25220010 | 0.01 |
ENSRNOT00000008786
|
Dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr1_+_104106245 | 0.01 |
ENSRNOT00000019175
ENSRNOT00000081660 |
Zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr6_+_12253788 | 0.01 |
ENSRNOT00000061675
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr5_-_162602339 | 0.01 |
ENSRNOT00000071779
|
RGD1563334
|
similar to novel protein similar to esterases |
| chr5_-_153840178 | 0.01 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr6_-_41842817 | 0.01 |
ENSRNOT00000084155
|
Lpin1
|
lipin 1 |
| chr14_+_33108024 | 0.01 |
ENSRNOT00000090350
ENSRNOT00000067650 ENSRNOT00000077242 |
Noa1
|
nitric oxide associated 1 |
| chr4_+_180062799 | 0.01 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr7_-_70661891 | 0.00 |
ENSRNOT00000010240
|
Inhbc
|
inhibin beta C subunit |
| chr9_+_61738471 | 0.00 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr1_-_80271001 | 0.00 |
ENSRNOT00000034266
|
Cd3eap
|
CD3e molecule associated protein |
| chr5_+_157820908 | 0.00 |
ENSRNOT00000068168
|
Emc1
|
ER membrane protein complex subunit 1 |
| chr6_-_79306443 | 0.00 |
ENSRNOT00000030706
|
Clec14a
|
C-type lectin domain family 14, member A |
| chr9_+_100956477 | 0.00 |
ENSRNOT00000078222
|
D2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr16_+_80729959 | 0.00 |
ENSRNOT00000082049
|
Tdrp
|
testis development related protein |
| chr8_+_119524039 | 0.00 |
ENSRNOT00000056050
|
Epm2aip1
|
EPM2A interacting protein 1 |
| chr18_+_70248713 | 0.00 |
ENSRNOT00000032202
|
Mbd1
|
methyl-CpG binding domain protein 1 |
| chr16_-_81880502 | 0.00 |
ENSRNOT00000079096
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr4_-_118487633 | 0.00 |
ENSRNOT00000068093
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr7_-_14369821 | 0.00 |
ENSRNOT00000008560
|
Akap8l
|
A-kinase anchoring protein 8 like |
| chrX_+_122507374 | 0.00 |
ENSRNOT00000032275
ENSRNOT00000080517 |
Dock11
|
dedicator of cytokinesis 11 |
| chr18_-_43945273 | 0.00 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr5_-_57683932 | 0.00 |
ENSRNOT00000074796
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chrX_+_76083549 | 0.00 |
ENSRNOT00000003573
|
Magee1
|
MAGE family member E1 |
| chr2_-_34923024 | 0.00 |
ENSRNOT00000090060
|
Cwc27
|
CWC27 spliceosome-associated protein homolog |
| chr9_+_69497121 | 0.00 |
ENSRNOT00000042562
|
Nrp2
|
neuropilin 2 |
| chr3_-_12007570 | 0.00 |
ENSRNOT00000060186
|
Lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr7_+_120273117 | 0.00 |
ENSRNOT00000014349
|
Galr3
|
galanin receptor 3 |
| chr1_-_82108083 | 0.00 |
ENSRNOT00000027677
ENSRNOT00000084530 |
Gsk3a
|
glycogen synthase kinase 3 alpha |
| chr2_+_140541619 | 0.00 |
ENSRNOT00000017396
|
Rab33b
|
RAB33B, member RAS oncogene family |
| chrX_-_2657155 | 0.00 |
ENSRNOT00000005630
|
Chst7
|
carbohydrate sulfotransferase 7 |
| chr10_-_74119009 | 0.00 |
ENSRNOT00000085712
ENSRNOT00000006926 |
Dhx40
|
DEAH-box helicase 40 |
| chr2_-_140464607 | 0.00 |
ENSRNOT00000058190
|
Ndufc1
|
NADH:ubiquinone oxidoreductase subunit C1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mga
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:0098749 | cerebellar neuron development(GO:0098749) |
| 0.0 | 0.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.0 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:2000820 | cardiac vascular smooth muscle cell development(GO:0060948) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.0 | 0.0 | GO:0015489 | ornithine decarboxylase inhibitor activity(GO:0008073) polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |