Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
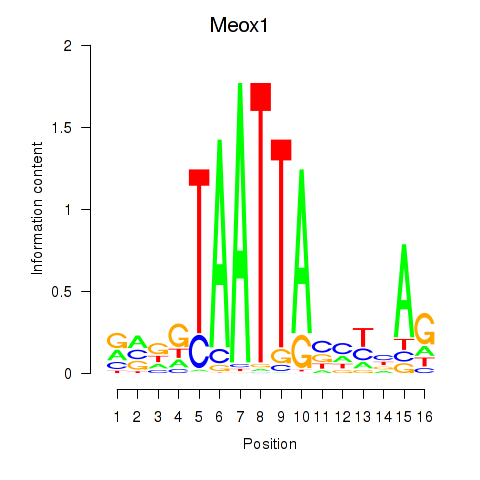
Results for Meox1
Z-value: 0.50
Transcription factors associated with Meox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox1
|
ENSRNOG00000020803 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox1 | rn6_v1_chr10_-_89816491_89816491 | 0.42 | 4.8e-01 | Click! |
Activity profile of Meox1 motif
Sorted Z-values of Meox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_123106694 | 0.71 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr5_+_50381244 | 0.48 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr1_+_87224677 | 0.32 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr1_+_80279706 | 0.16 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr7_-_143353925 | 0.16 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chr2_-_25235275 | 0.14 |
ENSRNOT00000061580
|
F2rl1
|
F2R like trypsin receptor 1 |
| chr6_+_34155017 | 0.14 |
ENSRNOT00000073663
|
Ttc32
|
tetratricopeptide repeat domain 32 |
| chr18_-_24735349 | 0.13 |
ENSRNOT00000036537
|
Gpr17
|
G protein-coupled receptor 17 |
| chr6_-_123577695 | 0.12 |
ENSRNOT00000006604
|
Foxn3
|
forkhead box N3 |
| chr20_+_40769586 | 0.12 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr10_-_44746549 | 0.11 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr8_-_78233430 | 0.11 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr4_-_157304653 | 0.11 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr20_-_10013190 | 0.11 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr9_+_20241062 | 0.11 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr12_+_47179664 | 0.11 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr12_-_13668515 | 0.10 |
ENSRNOT00000086847
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr17_-_2705123 | 0.09 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr14_-_34218961 | 0.08 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr20_+_31234493 | 0.08 |
ENSRNOT00000000675
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr5_+_76182121 | 0.08 |
ENSRNOT00000020707
|
Gng10
|
G protein subunit gamma 10 |
| chr8_+_126975833 | 0.08 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr3_-_154627257 | 0.08 |
ENSRNOT00000018328
|
Tgm2
|
transglutaminase 2 |
| chr2_+_158097843 | 0.08 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr10_-_93675991 | 0.08 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr20_-_30947484 | 0.07 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr1_-_154170409 | 0.07 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chrX_-_69218526 | 0.07 |
ENSRNOT00000092321
ENSRNOT00000074071 ENSRNOT00000092571 |
Pja1
|
praja ring finger ubiquitin ligase 1 |
| chr5_-_173233188 | 0.06 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr11_-_32550539 | 0.06 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chr1_-_15374850 | 0.06 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr3_+_63510293 | 0.06 |
ENSRNOT00000058093
|
Dfnb59
|
deafness, autosomal recessive 59 |
| chr20_-_10013559 | 0.06 |
ENSRNOT00000091623
|
Rsph1
|
radial spoke head 1 homolog |
| chrX_+_111396995 | 0.06 |
ENSRNOT00000087634
|
Pih1d3
|
PIH1 domain containing 3 |
| chr1_+_40879747 | 0.06 |
ENSRNOT00000083702
|
Akap12
|
A-kinase anchoring protein 12 |
| chr7_-_29171783 | 0.06 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr9_-_23493081 | 0.05 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr11_-_62067655 | 0.05 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_+_72395218 | 0.05 |
ENSRNOT00000057616
|
Prg3
|
proteoglycan 3, pro eosinophil major basic protein 2 |
| chr12_+_22165486 | 0.05 |
ENSRNOT00000001890
|
Mospd3
|
motile sperm domain containing 3 |
| chr2_+_185846232 | 0.05 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr6_+_22296128 | 0.05 |
ENSRNOT00000087805
|
Dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_+_89408935 | 0.05 |
ENSRNOT00000075068
|
LOC100909893
|
protein FAM187B-like |
| chr8_+_87205311 | 0.05 |
ENSRNOT00000014449
|
LOC100363289
|
LRRGT00022-like |
| chr1_+_248428099 | 0.05 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr18_-_67224566 | 0.04 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr12_-_9998779 | 0.04 |
ENSRNOT00000001265
|
Rpl21
|
ribosomal protein L21 |
| chr7_+_133856101 | 0.04 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr1_-_67065797 | 0.04 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr11_-_14304603 | 0.04 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr14_-_10694164 | 0.04 |
ENSRNOT00000080099
|
AC099384.2
|
|
| chr18_+_51523758 | 0.04 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr3_-_101547478 | 0.04 |
ENSRNOT00000006203
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr7_-_107203897 | 0.04 |
ENSRNOT00000086263
|
Lrrc6
|
leucine rich repeat containing 6 |
| chr5_+_15043955 | 0.04 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chrX_-_29556610 | 0.04 |
ENSRNOT00000068113
|
Trappc2
|
trafficking protein particle complex 2 |
| chr5_-_161981441 | 0.04 |
ENSRNOT00000020316
|
Pdpn
|
podoplanin |
| chr1_-_252461461 | 0.04 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr4_-_100252755 | 0.04 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr1_+_88113445 | 0.04 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr7_+_72924799 | 0.04 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr9_-_80167033 | 0.04 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr12_+_22727335 | 0.04 |
ENSRNOT00000077293
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr4_-_165026414 | 0.04 |
ENSRNOT00000071421
|
Klra1
|
killer cell lectin-like receptor, subfamily A, member 1 |
| chr19_-_49448072 | 0.03 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr4_+_78458625 | 0.03 |
ENSRNOT00000049891
|
Tmem176a
|
transmembrane protein 176A |
| chr7_+_2504695 | 0.03 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr7_-_145062956 | 0.03 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr7_-_143497108 | 0.03 |
ENSRNOT00000048613
|
Krt76
|
keratin 76 |
| chr8_+_103938520 | 0.03 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr9_+_92681078 | 0.03 |
ENSRNOT00000034152
|
Sp100
|
SP100 nuclear antigen |
| chr6_+_101532518 | 0.03 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr15_-_28517335 | 0.03 |
ENSRNOT00000060514
ENSRNOT00000090378 |
LOC100911576
|
heterogeneous nuclear ribonucleoproteins C1/C2-like |
| chrX_-_123980357 | 0.03 |
ENSRNOT00000049435
|
Rhox8
|
reproductive homeobox 8 |
| chr2_+_187951344 | 0.03 |
ENSRNOT00000027123
|
Ssr2
|
signal sequence receptor, beta |
| chr2_+_208900103 | 0.03 |
ENSRNOT00000055966
|
Chi3l3
|
chitinase 3-like 3 |
| chr11_+_70056624 | 0.03 |
ENSRNOT00000002447
|
AC133403.1
|
|
| chr7_+_120161091 | 0.03 |
ENSRNOT00000013764
|
Nol12
|
nucleolar protein 12 |
| chr15_+_27875911 | 0.03 |
ENSRNOT00000013582
|
Pnp
|
purine nucleoside phosphorylase |
| chr11_-_82893845 | 0.03 |
ENSRNOT00000075306
|
LOC100912571
|
eukaryotic translation initiation factor 4 gamma 1-like |
| chr14_+_88543630 | 0.03 |
ENSRNOT00000060515
|
Hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr6_+_132510757 | 0.03 |
ENSRNOT00000080230
|
Evl
|
Enah/Vasp-like |
| chr17_-_54678710 | 0.03 |
ENSRNOT00000046013
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr1_-_212633304 | 0.03 |
ENSRNOT00000025611
|
Olr286
|
olfactory receptor 286 |
| chr7_-_140356209 | 0.03 |
ENSRNOT00000077856
|
Rnd1
|
Rho family GTPase 1 |
| chr3_-_46942966 | 0.03 |
ENSRNOT00000087439
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr4_+_61924013 | 0.03 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr14_+_22517774 | 0.03 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr8_-_82533689 | 0.03 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr2_-_158133861 | 0.03 |
ENSRNOT00000090700
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_+_225037737 | 0.03 |
ENSRNOT00000077959
|
Bscl2
|
BSCL2, seipin lipid droplet biogenesis associated |
| chr4_+_55720010 | 0.03 |
ENSRNOT00000063987
|
Fscn3
|
fascin actin-bundling protein 3 |
| chr1_+_219390523 | 0.03 |
ENSRNOT00000054852
|
Gpr152
|
G protein-coupled receptor 152 |
| chr3_-_7498555 | 0.03 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr2_+_80269661 | 0.02 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr7_+_120153184 | 0.02 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr1_+_265157379 | 0.02 |
ENSRNOT00000022426
|
Btrc
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr8_+_26432514 | 0.02 |
ENSRNOT00000081574
|
Sept7
|
septin 7 |
| chr6_+_124123228 | 0.02 |
ENSRNOT00000005329
|
Psmc1
|
proteasome 26S subunit, ATPase 1 |
| chr7_-_59882077 | 0.02 |
ENSRNOT00000068774
|
Myrfl
|
myelin regulatory factor-like |
| chr1_+_69841998 | 0.02 |
ENSRNOT00000072231
|
Zfp773-ps1
|
zinc finger protein 773, pseudogene 1 |
| chr17_-_43537293 | 0.02 |
ENSRNOT00000091749
|
Slc17a3
|
solute carrier family 17 member 3 |
| chr6_-_127319362 | 0.02 |
ENSRNOT00000012256
|
Ddx24
|
DEAD-box helicase 24 |
| chr10_+_55492404 | 0.02 |
ENSRNOT00000005588
ENSRNOT00000078038 |
Rpl26
|
ribosomal protein L26 |
| chr3_-_52849907 | 0.02 |
ENSRNOT00000041096
|
Scn7a
|
sodium voltage-gated channel alpha subunit 7 |
| chr11_+_43194348 | 0.02 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr15_-_35113678 | 0.02 |
ENSRNOT00000059677
|
Ctsg
|
cathepsin G |
| chr15_+_4554603 | 0.02 |
ENSRNOT00000075153
|
Kcnk5
|
potassium two pore domain channel subfamily K member 5 |
| chr13_+_82438697 | 0.02 |
ENSRNOT00000003759
|
Selp
|
selectin P |
| chr10_+_103956019 | 0.02 |
ENSRNOT00000045875
|
Mrpl58
|
mitochondrial ribosomal protein L58 |
| chr14_-_64476796 | 0.02 |
ENSRNOT00000029104
|
Gba3
|
glucosidase, beta, acid 3 |
| chr11_-_69316848 | 0.02 |
ENSRNOT00000033420
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr4_+_70977556 | 0.02 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr7_+_144080614 | 0.02 |
ENSRNOT00000077687
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr2_+_22950018 | 0.02 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr1_+_169590308 | 0.02 |
ENSRNOT00000023147
|
Olr155
|
olfactory receptor 155 |
| chrX_-_70835089 | 0.02 |
ENSRNOT00000076351
|
Tex11
|
testis expressed 11 |
| chr2_+_178117466 | 0.02 |
ENSRNOT00000065115
ENSRNOT00000084198 |
LOC499643
|
similar to hypothetical protein FLJ25371 |
| chr2_-_60657712 | 0.02 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr9_-_44419998 | 0.02 |
ENSRNOT00000091397
ENSRNOT00000083747 |
Tsga10
|
testis specific 10 |
| chr19_-_24614019 | 0.02 |
ENSRNOT00000005124
|
Scoc
|
short coiled-coil protein |
| chr14_+_22375955 | 0.02 |
ENSRNOT00000063915
ENSRNOT00000034784 |
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr10_+_104582955 | 0.02 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr11_+_43143882 | 0.02 |
ENSRNOT00000041445
|
Olr1528
|
olfactory receptor 1528 |
| chr5_+_64476317 | 0.02 |
ENSRNOT00000017217
|
LOC108348074
|
collagen alpha-1(XV) chain-like |
| chr10_+_88227360 | 0.02 |
ENSRNOT00000041033
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr1_+_22332090 | 0.02 |
ENSRNOT00000091252
|
AABR07000663.1
|
trace amine-associated receptor 8c (Taar8c), mRNA |
| chr3_-_26056818 | 0.02 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr1_+_59156251 | 0.01 |
ENSRNOT00000017442
|
Lix1
|
limb and CNS expressed 1 |
| chr8_+_44136496 | 0.01 |
ENSRNOT00000087022
|
Scn3b
|
sodium voltage-gated channel beta subunit 3 |
| chr7_-_76294663 | 0.01 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr2_+_54466280 | 0.01 |
ENSRNOT00000033112
|
C6
|
complement C6 |
| chr4_+_115416580 | 0.01 |
ENSRNOT00000018303
|
Atp6v1b1
|
ATPase H+ transporting V1 subunit B1 |
| chr19_-_11263980 | 0.01 |
ENSRNOT00000025086
|
Nup93
|
nucleoporin 93 |
| chr4_+_181874861 | 0.01 |
ENSRNOT00000002527
|
Ccdc91
|
coiled-coil domain containing 91 |
| chr14_+_22091777 | 0.01 |
ENSRNOT00000072483
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr8_+_36850324 | 0.01 |
ENSRNOT00000060448
|
Pate2
|
prostate and testis expressed 2 |
| chr10_-_110274768 | 0.01 |
ENSRNOT00000054931
|
Sectm1a
|
secreted and transmembrane 1A |
| chr1_+_255579474 | 0.01 |
ENSRNOT00000024465
|
Btaf1
|
B-TFIID TATA-box binding protein associated factor 1 |
| chr2_-_219262901 | 0.01 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr9_+_71247781 | 0.01 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr18_-_69780922 | 0.01 |
ENSRNOT00000079093
|
Me2
|
malic enzyme 2 |
| chr17_-_44841382 | 0.01 |
ENSRNOT00000080119
|
Hist1h2ak
|
histone cluster 1, H2ak |
| chr1_-_103760913 | 0.01 |
ENSRNOT00000049722
|
Mrgprx2
|
MAS-related GPR, member X2 |
| chr5_-_157059508 | 0.01 |
ENSRNOT00000022282
|
Vwa5b1
|
von Willebrand factor A domain containing 5B1 |
| chr7_+_380741 | 0.01 |
ENSRNOT00000091013
|
LOC102554748
|
nidogen-2-like |
| chr18_+_4084228 | 0.01 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr5_-_102743417 | 0.01 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr1_+_273854248 | 0.01 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr3_-_114251647 | 0.01 |
ENSRNOT00000024245
|
Duoxa1
|
dual oxidase maturation factor 1 |
| chr13_-_89545182 | 0.01 |
ENSRNOT00000078402
|
Pcp4l1
|
Purkinje cell protein 4-like 1 |
| chr6_+_22167919 | 0.01 |
ENSRNOT00000007655
|
Nlrc4
|
NLR family, CARD domain containing 4 |
| chr6_-_78549669 | 0.01 |
ENSRNOT00000009940
|
Foxa1
|
forkhead box A1 |
| chr7_-_36660141 | 0.01 |
ENSRNOT00000012363
|
Nudt4
|
nudix hydrolase 4 |
| chr3_+_56056925 | 0.01 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chrX_+_6273733 | 0.01 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr8_+_22648323 | 0.01 |
ENSRNOT00000013165
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr17_+_82430836 | 0.01 |
ENSRNOT00000083164
|
Malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr20_-_3751994 | 0.01 |
ENSRNOT00000072288
|
Pou5f1
|
POU class 5 homeobox 1 |
| chrX_+_5825711 | 0.01 |
ENSRNOT00000037360
|
Efhc2
|
EF-hand domain containing 2 |
| chr1_+_227892956 | 0.01 |
ENSRNOT00000028483
|
AABR07006278.1
|
|
| chr3_-_72895740 | 0.01 |
ENSRNOT00000012568
|
Fads2l1
|
fatty acid desaturase 2-like 1 |
| chr8_-_87419564 | 0.01 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr16_-_74496731 | 0.01 |
ENSRNOT00000015790
|
Mrps31
|
mitochondrial ribosomal protein S31 |
| chr1_+_185863043 | 0.01 |
ENSRNOT00000079072
|
Sox6
|
SRY box 6 |
| chr11_+_88376256 | 0.01 |
ENSRNOT00000081776
|
Vpreb1
|
pre-B lymphocyte 1 |
| chr7_-_76488216 | 0.01 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr3_+_149802621 | 0.00 |
ENSRNOT00000071772
|
LOC690507
|
similar to Vomeromodulin |
| chr2_+_145174876 | 0.00 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr3_-_8348746 | 0.00 |
ENSRNOT00000064638
|
Trub2
|
TruB pseudouridine synthase family member 2 |
| chrX_+_66314418 | 0.00 |
ENSRNOT00000035405
|
Pgr15l
|
G protein-coupled receptor 15-like |
| chr1_-_23556241 | 0.00 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chr1_+_97777685 | 0.00 |
ENSRNOT00000030895
|
LOC690284
|
similar to F49E2.5d |
| chr9_+_98438439 | 0.00 |
ENSRNOT00000027220
|
Scly
|
selenocysteine lyase |
| chr3_-_21904133 | 0.00 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr13_+_47935163 | 0.00 |
ENSRNOT00000006768
|
Eif2d
|
eukaryotic translation initiation factor 2D |
| chr3_-_37803112 | 0.00 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr3_+_95233874 | 0.00 |
ENSRNOT00000079743
|
LOC691083
|
hypothetical protein LOC691083 |
| chr1_+_166141954 | 0.00 |
ENSRNOT00000042524
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr5_+_137257287 | 0.00 |
ENSRNOT00000037160
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr16_+_23553647 | 0.00 |
ENSRNOT00000041994
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_218810118 | 0.00 |
ENSRNOT00000065950
ENSRNOT00000020886 |
Ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr1_-_84950210 | 0.00 |
ENSRNOT00000083050
|
AABR07002784.1
|
|
| chr10_-_43931094 | 0.00 |
ENSRNOT00000039778
|
Olr1415
|
olfactory receptor 1415 |
| chr1_+_163398891 | 0.00 |
ENSRNOT00000020513
|
Gucy2e
|
guanylate cyclase 2E |
| chr14_-_82658891 | 0.00 |
ENSRNOT00000006841
|
Uvssa
|
UV-stimulated scaffold protein A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.5 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.0 | GO:1902276 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.0 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.0 | 0.0 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.0 | GO:0070638 | nicotinamide riboside catabolic process(GO:0006738) urate biosynthetic process(GO:0034418) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) purine-nucleoside phosphorylase activity(GO:0004731) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |