Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
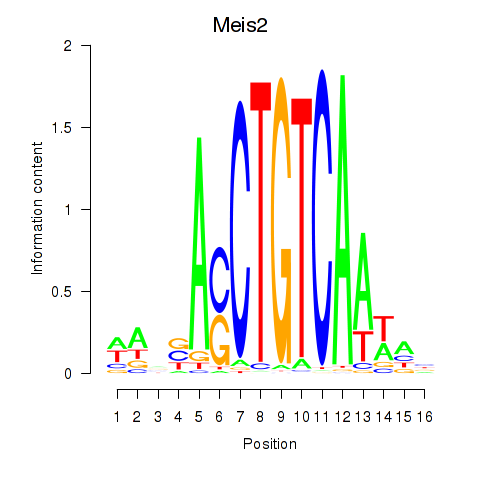
Results for Meis2
Z-value: 0.39
Transcription factors associated with Meis2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meis2
|
ENSRNOG00000004730 | Meis homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis2 | rn6_v1_chr3_-_107760550_107760589 | 0.94 | 1.8e-02 | Click! |
Activity profile of Meis2 motif
Sorted Z-values of Meis2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_52266100 | 0.27 |
ENSRNOT00000087532
|
AC096239.3
|
|
| chr11_-_88180542 | 0.17 |
ENSRNOT00000002539
|
Ypel1
|
yippee-like 1 |
| chr2_-_178521038 | 0.16 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr7_+_20462081 | 0.16 |
ENSRNOT00000088383
|
AABR07056131.1
|
|
| chr13_+_113373578 | 0.14 |
ENSRNOT00000009900
|
Plxna2
|
plexin A2 |
| chr5_+_8459660 | 0.13 |
ENSRNOT00000007491
|
Cpa6
|
carboxypeptidase A6 |
| chr5_+_118574801 | 0.12 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr11_+_45888221 | 0.11 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr13_-_82129989 | 0.11 |
ENSRNOT00000078963
|
AC124874.1
|
|
| chr7_+_20262680 | 0.11 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr9_-_73958480 | 0.11 |
ENSRNOT00000017838
|
Myl1
|
myosin, light chain 1 |
| chr1_+_220071811 | 0.11 |
ENSRNOT00000090642
|
AC126581.1
|
|
| chrX_+_45637415 | 0.10 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr7_-_70452675 | 0.10 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr3_-_102151489 | 0.08 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr9_-_49448168 | 0.08 |
ENSRNOT00000059478
|
AABR07067499.1
|
|
| chr10_-_110101872 | 0.08 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr3_-_52998650 | 0.08 |
ENSRNOT00000009988
|
RGD1560831
|
similar to 40S ribosomal protein S3 |
| chr2_-_19808937 | 0.08 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr4_+_181315444 | 0.08 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr3_-_162035614 | 0.08 |
ENSRNOT00000047890
|
Zfp663
|
zinc finger protein 663 |
| chr2_-_220838905 | 0.08 |
ENSRNOT00000078914
|
AABR07013065.1
|
|
| chr12_+_24148567 | 0.08 |
ENSRNOT00000077378
ENSRNOT00000045157 |
Ccl24
|
C-C motif chemokine ligand 24 |
| chr8_-_115358046 | 0.08 |
ENSRNOT00000017607
|
Grm2
|
glutamate metabotropic receptor 2 |
| chr4_-_182844291 | 0.07 |
ENSRNOT00000064320
|
Tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr11_-_81444375 | 0.07 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr13_+_101790865 | 0.07 |
ENSRNOT00000087784
|
Taf1a
|
TATA-box binding protein associated factor, RNA polymerase I subunit A |
| chr1_+_88875375 | 0.07 |
ENSRNOT00000028284
|
Tyrobp
|
Tyro protein tyrosine kinase binding protein |
| chr9_+_27343853 | 0.07 |
ENSRNOT00000072794
|
Tmem14a
|
transmembrane protein 14A |
| chr5_+_156613741 | 0.07 |
ENSRNOT00000076031
|
Sh2d5
|
SH2 domain containing 5 |
| chr18_+_47739140 | 0.07 |
ENSRNOT00000082760
|
Sncaip
|
synuclein, alpha interacting protein |
| chrX_-_10031167 | 0.07 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr5_+_147476221 | 0.07 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr5_-_105010857 | 0.07 |
ENSRNOT00000009749
|
Plin2
|
perilipin 2 |
| chr14_-_1461543 | 0.07 |
ENSRNOT00000075656
|
Crlf2
|
cytokine receptor-like factor 2 |
| chr16_+_39909270 | 0.07 |
ENSRNOT00000081994
|
Wdr17
|
WD repeat domain 17 |
| chrX_+_13117239 | 0.07 |
ENSRNOT00000088998
|
AABR07037112.1
|
|
| chr5_-_156105867 | 0.06 |
ENSRNOT00000078851
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr3_+_128155069 | 0.06 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr2_+_240021152 | 0.06 |
ENSRNOT00000012473
|
Tacr3
|
tachykinin receptor 3 |
| chr8_+_33239139 | 0.06 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr12_+_47074200 | 0.06 |
ENSRNOT00000014910
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr12_+_31530699 | 0.06 |
ENSRNOT00000033379
ENSRNOT00000067867 |
Rimbp2
|
RIMS binding protein 2 |
| chr8_+_119160731 | 0.06 |
ENSRNOT00000046745
|
Als2cl
|
ALS2 C-terminal like |
| chr10_-_76166251 | 0.06 |
ENSRNOT00000003251
ENSRNOT00000055683 |
Akap1
|
A-kinase anchoring protein 1 |
| chr1_-_82004538 | 0.06 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr11_+_84745904 | 0.06 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr5_-_2982603 | 0.06 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr8_-_14292698 | 0.06 |
ENSRNOT00000073672
|
AABR07069238.1
|
|
| chr2_-_104461863 | 0.06 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr5_-_152589719 | 0.06 |
ENSRNOT00000022522
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr20_+_28572242 | 0.06 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr9_+_61738471 | 0.06 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr8_-_73194837 | 0.06 |
ENSRNOT00000024885
ENSRNOT00000081450 ENSRNOT00000064613 |
Tln2
|
talin 2 |
| chr3_+_72329967 | 0.06 |
ENSRNOT00000090256
|
Slc43a3
|
solute carrier family 43, member 3 |
| chr3_-_122946289 | 0.05 |
ENSRNOT00000055810
|
Pced1a
|
PC-esterase domain containing 1A |
| chr7_-_15821927 | 0.05 |
ENSRNOT00000050658
|
LOC691422
|
similar to zinc finger protein 101 |
| chr1_+_15180328 | 0.05 |
ENSRNOT00000050656
|
Il20ra
|
interleukin 20 receptor subunit alpha |
| chr20_+_4576057 | 0.05 |
ENSRNOT00000081456
ENSRNOT00000085701 |
Ehmt2
|
euchromatic histone lysine methyltransferase 2 |
| chrX_+_72684329 | 0.05 |
ENSRNOT00000057644
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr5_-_74029238 | 0.05 |
ENSRNOT00000031432
|
Frrs1l
|
ferric-chelate reductase 1-like |
| chr3_-_153321352 | 0.05 |
ENSRNOT00000064824
|
Rbl1
|
RB transcriptional corepressor like 1 |
| chr1_+_91152635 | 0.05 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr19_-_55490426 | 0.05 |
ENSRNOT00000081800
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr17_-_77261731 | 0.05 |
ENSRNOT00000066850
|
Ucma
|
upper zone of growth plate and cartilage matrix associated |
| chr6_-_51297712 | 0.05 |
ENSRNOT00000082985
|
Prkar2b
|
protein kinase cAMP-dependent type 2 regulatory subunit beta |
| chr18_+_56414488 | 0.05 |
ENSRNOT00000088988
|
Csf1r
|
colony stimulating factor 1 receptor |
| chr1_+_87045796 | 0.05 |
ENSRNOT00000027620
|
Lgals7
|
galectin 7 |
| chr6_+_59757397 | 0.05 |
ENSRNOT00000047001
|
AABR07064000.1
|
|
| chr14_+_82350734 | 0.05 |
ENSRNOT00000023259
|
Tmem129
|
transmembrane protein 129 |
| chr14_-_78377825 | 0.05 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr10_-_110585376 | 0.05 |
ENSRNOT00000054917
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr12_-_6341902 | 0.05 |
ENSRNOT00000001201
|
Hsph1
|
heat shock protein family H (Hsp110) member 1 |
| chr14_+_104250617 | 0.05 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_-_14369821 | 0.05 |
ENSRNOT00000008560
|
Akap8l
|
A-kinase anchoring protein 8 like |
| chr7_-_120744602 | 0.05 |
ENSRNOT00000018564
|
Kcnj4
|
potassium voltage-gated channel subfamily J member 4 |
| chr2_-_149432106 | 0.05 |
ENSRNOT00000051027
|
P2ry13
|
purinergic receptor P2Y13 |
| chr2_-_186232292 | 0.05 |
ENSRNOT00000087088
|
Dclk2
|
doublecortin-like kinase 2 |
| chr1_+_248195797 | 0.05 |
ENSRNOT00000066891
|
LOC103690131
|
tumor protein D55-like |
| chr1_+_166534643 | 0.04 |
ENSRNOT00000026586
|
Pde2a
|
phosphodiesterase 2A |
| chr3_+_80555196 | 0.04 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chrX_-_23187341 | 0.04 |
ENSRNOT00000000180
|
Alas2
|
5'-aminolevulinate synthase 2 |
| chr1_-_82003691 | 0.04 |
ENSRNOT00000084569
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr3_+_58632476 | 0.04 |
ENSRNOT00000010630
|
Rapgef4
|
Rap guanine nucleotide exchange factor 4 |
| chrX_+_15114892 | 0.04 |
ENSRNOT00000078168
|
Wdr13
|
WD repeat domain 13 |
| chr10_+_36715565 | 0.04 |
ENSRNOT00000005048
|
Clk4
|
CDC-like kinase 4 |
| chr10_+_105498728 | 0.04 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr1_-_48686713 | 0.04 |
ENSRNOT00000091196
|
AABR07001516.1
|
|
| chr14_-_34115273 | 0.04 |
ENSRNOT00000032156
|
Cep135
|
centrosomal protein 135 |
| chr6_-_94834908 | 0.04 |
ENSRNOT00000006284
|
L3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr10_+_92667869 | 0.04 |
ENSRNOT00000082780
ENSRNOT00000073350 |
Itgb3
|
integrin subunit beta 3 |
| chr5_+_50381244 | 0.04 |
ENSRNOT00000012385
|
Cga
|
glycoprotein hormones, alpha polypeptide |
| chr7_+_70697050 | 0.04 |
ENSRNOT00000045964
|
R3hdm2
|
R3H domain containing 2 |
| chr17_-_43626538 | 0.04 |
ENSRNOT00000074292
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr11_-_38420032 | 0.04 |
ENSRNOT00000002209
ENSRNOT00000080681 |
C2cd2
|
C2 calcium-dependent domain containing 2 |
| chr10_+_62191656 | 0.04 |
ENSRNOT00000029866
|
Smyd4
|
SET and MYND domain containing 4 |
| chr4_-_109532234 | 0.04 |
ENSRNOT00000008665
|
Reg3g
|
regenerating family member 3 gamma |
| chr2_-_219628997 | 0.04 |
ENSRNOT00000064484
|
Trmt13
|
tRNA methyltransferase 13 homolog |
| chr5_-_48165317 | 0.04 |
ENSRNOT00000088179
|
Ankrd6
|
ankyrin repeat domain 6 |
| chr15_-_103340239 | 0.04 |
ENSRNOT00000067309
ENSRNOT00000088909 |
Tgds
|
TDP-glucose 4,6-dehydratase |
| chr19_-_50220455 | 0.04 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr19_+_14459450 | 0.04 |
ENSRNOT00000079257
|
Zfp82
|
zinc finger protein 82 |
| chr1_+_191704311 | 0.04 |
ENSRNOT00000024057
|
Scnn1g
|
sodium channel epithelial 1 gamma subunit |
| chr1_-_78626284 | 0.04 |
ENSRNOT00000052299
|
LOC108349752
|
carcinoembryonic antigen-related cell adhesion molecule 3-like |
| chr4_-_119568736 | 0.04 |
ENSRNOT00000041234
|
Aplf
|
aprataxin and PNKP like factor |
| chr11_+_46737892 | 0.04 |
ENSRNOT00000052182
|
AABR07033979.1
|
|
| chr16_-_6245644 | 0.04 |
ENSRNOT00000040759
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr7_+_123482255 | 0.04 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr1_+_141218095 | 0.04 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr19_+_25143428 | 0.04 |
ENSRNOT00000007535
|
Palm3
|
paralemmin 3 |
| chr7_-_3170146 | 0.04 |
ENSRNOT00000036373
|
Dgka
|
diacylglycerol kinase, alpha |
| chr13_+_104816371 | 0.04 |
ENSRNOT00000086497
|
AABR07022057.1
|
|
| chr13_-_37287458 | 0.04 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr17_-_14058642 | 0.04 |
ENSRNOT00000019489
|
Nxnl2
|
nucleoredoxin-like 2 |
| chr5_-_173680290 | 0.04 |
ENSRNOT00000027564
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr12_+_41200718 | 0.04 |
ENSRNOT00000038426
ENSRNOT00000048450 ENSRNOT00000067176 |
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr10_-_58834538 | 0.04 |
ENSRNOT00000020043
|
Slc13a5
|
solute carrier family 13 member 5 |
| chr11_+_80358211 | 0.04 |
ENSRNOT00000002519
|
Sst
|
somatostatin |
| chr17_-_43675934 | 0.04 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr12_+_660011 | 0.04 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr9_-_40008680 | 0.04 |
ENSRNOT00000016578
|
Khdrbs2
|
KH RNA binding domain containing, signal transduction associated 2 |
| chr3_+_36173002 | 0.04 |
ENSRNOT00000038906
|
LOC499796
|
LRRGT00012 |
| chr14_-_5428757 | 0.04 |
ENSRNOT00000002897
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr3_-_123997277 | 0.04 |
ENSRNOT00000058238
|
RGD1564425
|
similar to Proteasome subunit beta type 3 (Proteasome theta chain) |
| chr16_+_7336685 | 0.04 |
ENSRNOT00000025853
|
Bap1
|
Brca1 associated protein 1 |
| chr8_-_39551700 | 0.04 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr6_-_132802206 | 0.03 |
ENSRNOT00000080163
ENSRNOT00000050350 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr6_+_6908684 | 0.03 |
ENSRNOT00000079095
|
Mta3
|
metastasis associated 1 family, member 3 |
| chr1_-_88112683 | 0.03 |
ENSRNOT00000090615
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr13_-_83202864 | 0.03 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr8_+_56179816 | 0.03 |
ENSRNOT00000059078
|
Arhgap20
|
Rho GTPase activating protein 20 |
| chr7_-_57679795 | 0.03 |
ENSRNOT00000007461
|
Trhde
|
thyrotropin-releasing hormone degrading enzyme |
| chr4_-_116786391 | 0.03 |
ENSRNOT00000086297
ENSRNOT00000091490 |
Exoc6b
|
exocyst complex component 6B |
| chr1_-_199655147 | 0.03 |
ENSRNOT00000026979
|
LOC103691238
|
zinc finger protein 239-like |
| chr8_-_55696601 | 0.03 |
ENSRNOT00000016127
|
RGD1562914
|
RGD1562914 |
| chr4_+_88423412 | 0.03 |
ENSRNOT00000040163
|
Vom1r88
|
vomeronasal 1 receptor 88 |
| chr17_+_2690062 | 0.03 |
ENSRNOT00000024937
|
Olr1653
|
olfactory receptor 1653 |
| chr2_+_147006830 | 0.03 |
ENSRNOT00000080780
|
AABR07010672.1
|
|
| chr1_+_72756809 | 0.03 |
ENSRNOT00000082423
|
Ppp6r1
|
protein phosphatase 6, regulatory subunit 1 |
| chr5_+_145079803 | 0.03 |
ENSRNOT00000084202
|
Sfpq
|
splicing factor proline and glutamine rich |
| chr2_+_183674522 | 0.03 |
ENSRNOT00000014433
|
Tmem154
|
transmembrane protein 154 |
| chr4_+_22859622 | 0.03 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chrX_-_45419817 | 0.03 |
ENSRNOT00000087509
|
Ptges3l1
|
prostaglandin E synthase 3-like 1 |
| chr18_+_30387937 | 0.03 |
ENSRNOT00000027210
|
Pcdhb4
|
protocadherin beta 4 |
| chr8_+_119228612 | 0.03 |
ENSRNOT00000078439
ENSRNOT00000043737 |
Lrrc2
|
leucine rich repeat containing 2 |
| chr7_-_107223047 | 0.03 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr6_-_108976489 | 0.03 |
ENSRNOT00000007350
|
Rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr2_+_211337271 | 0.03 |
ENSRNOT00000045155
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr8_-_62616828 | 0.03 |
ENSRNOT00000068340
|
Arid3b
|
AT-rich interaction domain 3B |
| chr5_+_149047681 | 0.03 |
ENSRNOT00000015198
|
Laptm5
|
lysosomal protein transmembrane 5 |
| chr4_+_81311490 | 0.03 |
ENSRNOT00000016265
|
Snx10
|
sorting nexin 10 |
| chr17_+_87274944 | 0.03 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr10_-_11935382 | 0.03 |
ENSRNOT00000010059
|
Zfp174
|
zinc finger protein 174 |
| chrX_+_77143892 | 0.03 |
ENSRNOT00000085227
|
Atp7a
|
ATPase copper transporting alpha |
| chr3_-_160573978 | 0.03 |
ENSRNOT00000018386
|
Wfdc5
|
WAP four-disulfide core domain 5 |
| chr2_-_227411964 | 0.03 |
ENSRNOT00000019931
|
Synpo2
|
synaptopodin 2 |
| chr3_-_51297852 | 0.03 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr15_-_70399924 | 0.03 |
ENSRNOT00000087940
|
Diaph3
|
diaphanous-related formin 3 |
| chr1_-_142615673 | 0.03 |
ENSRNOT00000018021
|
Iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr5_-_59543101 | 0.03 |
ENSRNOT00000019532
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr3_+_7686503 | 0.03 |
ENSRNOT00000017984
|
Setx
|
senataxin |
| chr1_-_217620420 | 0.03 |
ENSRNOT00000092783
|
Cttn
|
cortactin |
| chr8_-_107656861 | 0.03 |
ENSRNOT00000050374
|
Mras
|
muscle RAS oncogene homolog |
| chrX_+_984798 | 0.03 |
ENSRNOT00000073016
|
Zfp182
|
zinc finger protein 182 |
| chr3_+_156642464 | 0.03 |
ENSRNOT00000085354
|
Top1
|
topoisomerase (DNA) I |
| chr5_+_145145983 | 0.03 |
ENSRNOT00000079732
|
Zmym6
|
zinc finger MYM-type containing 6 |
| chr1_-_195096694 | 0.03 |
ENSRNOT00000088874
|
Snurf
|
SNRPN upstream reading frame |
| chr1_+_61314070 | 0.03 |
ENSRNOT00000077642
|
Zfp54
|
zinc finger protein 54 |
| chr1_-_261051498 | 0.03 |
ENSRNOT00000071417
|
Arhgap19
|
Rho GTPase activating protein 19 |
| chr2_-_173542284 | 0.03 |
ENSRNOT00000013111
|
Zbbx
|
zinc finger, B-box domain containing |
| chr20_-_30748784 | 0.03 |
ENSRNOT00000084391
|
Sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr3_+_100768637 | 0.03 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr8_-_33121002 | 0.03 |
ENSRNOT00000047211
|
LOC100362981
|
LRRGT00010-like |
| chr5_-_57008795 | 0.03 |
ENSRNOT00000090891
ENSRNOT00000046463 |
Aptx
|
aprataxin |
| chr7_-_34951644 | 0.03 |
ENSRNOT00000030015
|
Vezt
|
vezatin, adherens junctions transmembrane protein |
| chr14_+_17531398 | 0.03 |
ENSRNOT00000050038
|
Cdkl2
|
cyclin dependent kinase like 2 |
| chr1_-_81295442 | 0.03 |
ENSRNOT00000030900
|
Irgc
|
immunity related GTPase cinema |
| chr7_-_8060373 | 0.03 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chr10_+_15183803 | 0.03 |
ENSRNOT00000039777
|
Fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr7_+_143874734 | 0.03 |
ENSRNOT00000093277
|
Gm9918
|
predicted gene 9918 |
| chr4_+_145653602 | 0.03 |
ENSRNOT00000038964
|
Tatdn2
|
TatD DNase domain containing 2 |
| chr5_+_56797502 | 0.03 |
ENSRNOT00000067992
|
Tmem215
|
transmembrane protein 215 |
| chr7_-_105592804 | 0.03 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr4_+_120671489 | 0.03 |
ENSRNOT00000029125
|
Mgll
|
monoglyceride lipase |
| chr3_-_113231790 | 0.03 |
ENSRNOT00000019025
|
Tp53bp1
|
tumor protein p53 binding protein 1 |
| chr3_+_28627084 | 0.03 |
ENSRNOT00000049884
|
Arhgap15
|
Rho GTPase activating protein 15 |
| chr6_-_55001464 | 0.03 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr16_-_71237118 | 0.03 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr9_+_81518176 | 0.02 |
ENSRNOT00000078317
ENSRNOT00000019265 ENSRNOT00000088246 ENSRNOT00000084682 |
Arpc2
|
actin related protein 2/3 complex, subunit 2 |
| chr15_-_51967806 | 0.02 |
ENSRNOT00000012993
ENSRNOT00000084166 ENSRNOT00000091826 ENSRNOT00000082505 |
Ppp3cc
|
protein phosphatase 3 catalytic subunit gamma |
| chr11_+_38727048 | 0.02 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr1_-_116567189 | 0.02 |
ENSRNOT00000048996
|
LOC100912195
|
protein BEX1-like |
| chr7_-_115045802 | 0.02 |
ENSRNOT00000040236
ENSRNOT00000071818 |
Mroh5
|
maestro heat-like repeat family member 5 |
| chr1_+_161922141 | 0.02 |
ENSRNOT00000078300
ENSRNOT00000015784 |
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr11_+_34557679 | 0.02 |
ENSRNOT00000002289
|
Ripply3
|
ripply transcriptional repressor 3 |
| chr2_-_259150479 | 0.02 |
ENSRNOT00000085892
|
AABR07013843.1
|
|
| chrX_+_28072826 | 0.02 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr3_+_150574473 | 0.02 |
ENSRNOT00000023905
|
Asip
|
agouti signaling protein |
| chr17_+_76306585 | 0.02 |
ENSRNOT00000065978
|
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr3_-_147143576 | 0.02 |
ENSRNOT00000091811
ENSRNOT00000012727 |
Snph
|
syntaphilin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Meis2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.0 | 0.1 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0071350 | monocyte extravasation(GO:0035696) interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.0 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.0 | GO:2000474 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.0 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.0 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.1 | GO:0097338 | response to antipsychotic drug(GO:0097332) response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.0 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.0 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |