Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Meis1
Z-value: 0.41
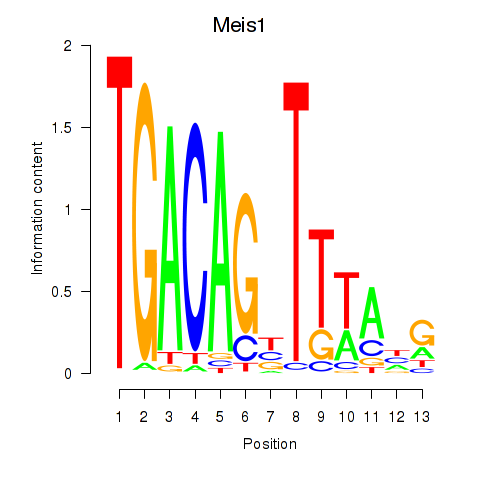
Transcription factors associated with Meis1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meis1
|
ENSRNOG00000004606 | Meis homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meis1 | rn6_v1_chr14_-_103321270_103321270 | -0.27 | 6.6e-01 | Click! |
Activity profile of Meis1 motif
Sorted Z-values of Meis1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_43357888 | 0.48 |
ENSRNOT00000022329
|
Scgn
|
secretagogin, EF-hand calcium binding protein |
| chr3_-_165039707 | 0.15 |
ENSRNOT00000081648
|
Kcng1
|
potassium voltage-gated channel modifier subfamily G member 1 |
| chr3_+_64190973 | 0.15 |
ENSRNOT00000011342
|
AABR07052587.1
|
|
| chr7_+_133856101 | 0.13 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr7_-_124929025 | 0.12 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr7_-_95669809 | 0.12 |
ENSRNOT00000006423
|
Sntb1
|
syntrophin, beta 1 |
| chr7_+_20462081 | 0.11 |
ENSRNOT00000088383
|
AABR07056131.1
|
|
| chr17_-_51912496 | 0.11 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr17_-_89163113 | 0.11 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr1_-_166912524 | 0.10 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr8_+_49713190 | 0.09 |
ENSRNOT00000022074
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2 |
| chr7_+_73222730 | 0.08 |
ENSRNOT00000007816
|
Erich5
|
glutamate-rich 5 |
| chr7_+_124680294 | 0.08 |
ENSRNOT00000014720
|
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr20_-_30947484 | 0.08 |
ENSRNOT00000065614
|
Pald1
|
phosphatase domain containing, paladin 1 |
| chr2_+_240021152 | 0.08 |
ENSRNOT00000012473
|
Tacr3
|
tachykinin receptor 3 |
| chr11_-_32508420 | 0.08 |
ENSRNOT00000002717
|
Kcne1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr16_+_21061237 | 0.08 |
ENSRNOT00000092136
|
Ncan
|
neurocan |
| chr18_+_56414488 | 0.08 |
ENSRNOT00000088988
|
Csf1r
|
colony stimulating factor 1 receptor |
| chrX_-_152642531 | 0.07 |
ENSRNOT00000085037
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr7_-_140437467 | 0.07 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr7_-_119071712 | 0.07 |
ENSRNOT00000037611
|
Myh9l1
|
myosin heavy chain 9-like 1 |
| chr2_-_211322719 | 0.07 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr13_-_70625842 | 0.07 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr14_+_104250617 | 0.06 |
ENSRNOT00000079874
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr2_-_219628997 | 0.06 |
ENSRNOT00000064484
|
Trmt13
|
tRNA methyltransferase 13 homolog |
| chr3_-_109044420 | 0.06 |
ENSRNOT00000007687
|
Rasgrp1
|
RAS guanyl releasing protein 1 |
| chr7_-_68512397 | 0.06 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr16_-_6245644 | 0.06 |
ENSRNOT00000040759
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chrX_+_28072826 | 0.06 |
ENSRNOT00000039796
|
Frmpd4
|
FERM and PDZ domain containing 4 |
| chr7_-_114590119 | 0.06 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr13_+_50091430 | 0.06 |
ENSRNOT00000046273
|
Atp2b4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr7_-_117680004 | 0.06 |
ENSRNOT00000040422
|
Slc39a4
|
solute carrier family 39 member 4 |
| chr1_-_199655147 | 0.06 |
ENSRNOT00000026979
|
LOC103691238
|
zinc finger protein 239-like |
| chr3_-_162035614 | 0.06 |
ENSRNOT00000047890
|
Zfp663
|
zinc finger protein 663 |
| chr11_-_64952687 | 0.06 |
ENSRNOT00000087892
|
Popdc2
|
popeye domain containing 2 |
| chr3_-_105214989 | 0.06 |
ENSRNOT00000037895
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr15_+_96821832 | 0.06 |
ENSRNOT00000012785
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr1_+_212181374 | 0.06 |
ENSRNOT00000085921
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr14_-_16903242 | 0.05 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr19_+_38768467 | 0.05 |
ENSRNOT00000027346
|
Cdh1
|
cadherin 1 |
| chr5_+_145257714 | 0.05 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr9_+_60021534 | 0.05 |
ENSRNOT00000093748
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr1_+_15180328 | 0.05 |
ENSRNOT00000050656
|
Il20ra
|
interleukin 20 receptor subunit alpha |
| chr5_+_147476221 | 0.05 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr7_-_105592804 | 0.05 |
ENSRNOT00000006789
|
Adcy8
|
adenylate cyclase 8 |
| chr7_-_69104950 | 0.05 |
ENSRNOT00000049063
|
LOC500846
|
hypothetical protein LOC500846 |
| chr10_-_55997299 | 0.05 |
ENSRNOT00000074505
|
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr10_-_4249739 | 0.05 |
ENSRNOT00000003150
|
Snx29
|
sorting nexin 29 |
| chr13_-_37287458 | 0.05 |
ENSRNOT00000003391
|
Insig2
|
insulin induced gene 2 |
| chr12_+_14021727 | 0.05 |
ENSRNOT00000060608
|
Mmd2
|
monocyte to macrophage differentiation-associated 2 |
| chr8_-_123371257 | 0.05 |
ENSRNOT00000017243
|
Stt3b
|
STT3B, catalytic subunit of the oligosaccharyltransferase complex |
| chr5_+_74727494 | 0.05 |
ENSRNOT00000076683
|
Rn50_5_0791.1
|
|
| chr6_+_108820774 | 0.05 |
ENSRNOT00000089683
ENSRNOT00000080038 ENSRNOT00000006492 |
Ylpm1
|
YLP motif containing 1 |
| chr1_+_199720038 | 0.05 |
ENSRNOT00000027297
|
Ahsp
|
alpha hemoglobin stabilizing protein |
| chr12_+_660011 | 0.05 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr9_-_119818310 | 0.05 |
ENSRNOT00000019359
|
Smchd1
|
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr17_-_10952441 | 0.05 |
ENSRNOT00000024580
|
Hrh2
|
histamine receptor H 2 |
| chr2_+_11658568 | 0.05 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr13_-_75059326 | 0.05 |
ENSRNOT00000089062
|
Rasal2
|
RAS protein activator like 2 |
| chr8_-_53816447 | 0.04 |
ENSRNOT00000011454
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr5_+_141530211 | 0.04 |
ENSRNOT00000056546
|
4933427I04Rik
|
Riken cDNA 4933427I04 gene |
| chr4_+_181315444 | 0.04 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr1_+_267618565 | 0.04 |
ENSRNOT00000076251
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr14_-_114692764 | 0.04 |
ENSRNOT00000008210
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr14_-_34115273 | 0.04 |
ENSRNOT00000032156
|
Cep135
|
centrosomal protein 135 |
| chr18_+_83777665 | 0.04 |
ENSRNOT00000018682
|
Cbln2
|
cerebellin 2 precursor |
| chrX_+_32495809 | 0.04 |
ENSRNOT00000020999
|
RGD1565844
|
similar to RIKEN cDNA 1700045I19 |
| chr10_-_25890639 | 0.04 |
ENSRNOT00000085499
|
Hmmr
|
hyaluronan-mediated motility receptor |
| chr4_-_118342176 | 0.04 |
ENSRNOT00000032477
|
AC139642.2
|
|
| chr2_-_199354793 | 0.04 |
ENSRNOT00000023548
|
Bcl9
|
B-cell CLL/lymphoma 9 |
| chr8_+_129617812 | 0.04 |
ENSRNOT00000079085
|
Ctnnb1
|
catenin beta 1 |
| chr17_+_76306585 | 0.04 |
ENSRNOT00000065978
|
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr7_+_11930135 | 0.04 |
ENSRNOT00000025391
|
Btbd2
|
BTB domain containing 2 |
| chr7_-_122340943 | 0.04 |
ENSRNOT00000078345
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr6_-_138249382 | 0.04 |
ENSRNOT00000085678
ENSRNOT00000006912 |
Ighm
|
immunoglobulin heavy constant mu |
| chr3_+_152626757 | 0.04 |
ENSRNOT00000086425
|
Aar2
|
AAR2 splicing factor homolog |
| chr4_+_149261044 | 0.04 |
ENSRNOT00000066670
|
Cxcl12
|
C-X-C motif chemokine ligand 12 |
| chr4_-_62840357 | 0.04 |
ENSRNOT00000059892
|
Slc13a4
|
solute carrier family 13 member 4 |
| chr17_-_84247038 | 0.04 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr18_+_65285318 | 0.04 |
ENSRNOT00000020431
|
Tcf4
|
transcription factor 4 |
| chr20_+_7330250 | 0.04 |
ENSRNOT00000090993
|
LOC499407
|
LRRGT00097 |
| chr7_-_75422268 | 0.04 |
ENSRNOT00000080218
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1 |
| chr17_-_10818835 | 0.04 |
ENSRNOT00000091046
|
Cplx2
|
complexin 2 |
| chr18_-_70184337 | 0.04 |
ENSRNOT00000020637
|
Ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr14_-_80130139 | 0.04 |
ENSRNOT00000091652
ENSRNOT00000010482 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr20_-_5166448 | 0.04 |
ENSRNOT00000076331
|
Aif1
|
allograft inflammatory factor 1 |
| chr20_-_5166252 | 0.04 |
ENSRNOT00000001138
|
Aif1
|
allograft inflammatory factor 1 |
| chr3_-_51297852 | 0.03 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr5_-_128009735 | 0.03 |
ENSRNOT00000013913
|
Gpx7
|
glutathione peroxidase 7 |
| chr1_-_44474674 | 0.03 |
ENSRNOT00000078768
|
Tfb1m
|
transcription factor B1, mitochondrial |
| chr2_-_125323454 | 0.03 |
ENSRNOT00000045837
|
Ankrd50
|
ankyrin repeat domain 50 |
| chr17_-_27602934 | 0.03 |
ENSRNOT00000079298
|
Rreb1
|
ras responsive element binding protein 1 |
| chr6_-_76535517 | 0.03 |
ENSRNOT00000083857
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr14_-_28967980 | 0.03 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr19_-_50220455 | 0.03 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr7_-_124009195 | 0.03 |
ENSRNOT00000082524
|
Poldip3
|
DNA polymerase delta interacting protein 3 |
| chr20_-_32139789 | 0.03 |
ENSRNOT00000078140
|
Srgn
|
serglycin |
| chr2_-_232245319 | 0.03 |
ENSRNOT00000014510
|
LOC691931
|
hypothetical protein LOC691931 |
| chr11_+_73738433 | 0.03 |
ENSRNOT00000002353
|
Tmem44
|
transmembrane protein 44 |
| chr2_+_143475323 | 0.03 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr10_+_69434941 | 0.03 |
ENSRNOT00000009756
|
Ccl11
|
C-C motif chemokine ligand 11 |
| chr2_-_123851854 | 0.03 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr8_-_63750531 | 0.03 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr3_+_138974871 | 0.03 |
ENSRNOT00000012524
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr3_-_150960030 | 0.03 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chr11_-_85895613 | 0.03 |
ENSRNOT00000071462
|
LOC100911837
|
protein HIRA-like |
| chr19_-_30821999 | 0.03 |
ENSRNOT00000072648
|
Gm8797
|
predicted pseudogene 8797 |
| chr13_-_48284408 | 0.03 |
ENSRNOT00000085967
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr20_-_32133431 | 0.03 |
ENSRNOT00000000443
|
Srgn
|
serglycin |
| chr3_-_12502859 | 0.03 |
ENSRNOT00000022814
|
Zbtb43
|
zinc finger and BTB domain containing 43 |
| chr20_-_4542073 | 0.03 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chrX_-_136150209 | 0.03 |
ENSRNOT00000048498
|
Enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr14_-_17616170 | 0.03 |
ENSRNOT00000090938
|
Thap6
|
THAP domain containing 6 |
| chr3_+_79823945 | 0.03 |
ENSRNOT00000014484
|
Celf1
|
CUGBP, Elav-like family member 1 |
| chrX_+_131381134 | 0.03 |
ENSRNOT00000007474
|
AABR07041481.1
|
|
| chr10_+_46722109 | 0.03 |
ENSRNOT00000032685
|
Drc3
|
dynein regulatory complex subunit 3 |
| chrX_+_43881246 | 0.03 |
ENSRNOT00000005153
|
LOC317456
|
hypothetical LOC317456 |
| chr9_+_66495832 | 0.03 |
ENSRNOT00000022676
ENSRNOT00000091283 |
Nop58
|
NOP58 ribonucleoprotein |
| chr18_-_67224566 | 0.03 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr7_+_11769400 | 0.03 |
ENSRNOT00000044417
|
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr10_+_53570989 | 0.02 |
ENSRNOT00000064764
ENSRNOT00000004516 |
Tmem220
|
transmembrane protein 220 |
| chr16_-_71237118 | 0.02 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr8_-_39190457 | 0.02 |
ENSRNOT00000090161
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr11_-_67702955 | 0.02 |
ENSRNOT00000084540
ENSRNOT00000078285 |
Kpna1
|
karyopherin subunit alpha 1 |
| chr4_-_117607428 | 0.02 |
ENSRNOT00000021243
|
LOC103690139
|
probable N-acetyltransferase CML6 |
| chr18_+_30114708 | 0.02 |
ENSRNOT00000027343
|
Pcdha4
|
protocadherin alpha 4 |
| chr1_+_105113595 | 0.02 |
ENSRNOT00000020853
ENSRNOT00000079655 |
Prmt3
|
protein arginine methyltransferase 3 |
| chr19_-_10826895 | 0.02 |
ENSRNOT00000090217
|
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr20_+_4392626 | 0.02 |
ENSRNOT00000000495
|
Prrt1
|
proline-rich transmembrane protein 1 |
| chr2_+_220432037 | 0.02 |
ENSRNOT00000021988
|
Frrs1
|
ferric-chelate reductase 1 |
| chr12_-_41625403 | 0.02 |
ENSRNOT00000001876
|
Sds
|
serine dehydratase |
| chr6_+_21051327 | 0.02 |
ENSRNOT00000052131
|
Fam98a
|
family with sequence similarity 98, member A |
| chr9_+_11064605 | 0.02 |
ENSRNOT00000075121
|
Stap2
|
signal transducing adaptor family member 2 |
| chr19_-_55423052 | 0.02 |
ENSRNOT00000019528
ENSRNOT00000079641 |
Galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr1_+_87650207 | 0.02 |
ENSRNOT00000077161
|
Zfp84
|
zinc finger protein 84 |
| chr1_+_265829108 | 0.02 |
ENSRNOT00000025791
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chrX_+_106390759 | 0.02 |
ENSRNOT00000049375
|
Bhlhb9
|
basic helix-loop-helix domain containing, class B, 9 |
| chr10_+_45966926 | 0.02 |
ENSRNOT00000067584
|
Olfr225
|
olfactory receptor 225 |
| chr2_-_80667481 | 0.02 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr19_-_25961666 | 0.02 |
ENSRNOT00000004091
|
Calr
|
calreticulin |
| chr5_-_173680290 | 0.02 |
ENSRNOT00000027564
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr15_+_52234563 | 0.02 |
ENSRNOT00000015169
|
Reep4
|
receptor accessory protein 4 |
| chr7_+_13318533 | 0.02 |
ENSRNOT00000034545
|
LOC691170
|
similar to zinc finger protein 84 (HPF2) |
| chr13_+_52889737 | 0.02 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr11_-_65350442 | 0.02 |
ENSRNOT00000003773
|
Gpr156
|
G protein-coupled receptor 156 |
| chr1_+_141218095 | 0.02 |
ENSRNOT00000051411
|
LOC691427
|
similar to 6.8 kDa mitochondrial proteolipid |
| chr19_-_37667987 | 0.02 |
ENSRNOT00000059577
|
Acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr15_-_35113678 | 0.02 |
ENSRNOT00000059677
|
Ctsg
|
cathepsin G |
| chr19_+_60017746 | 0.02 |
ENSRNOT00000042623
|
Pard3
|
par-3 family cell polarity regulator |
| chr16_+_19792838 | 0.02 |
ENSRNOT00000022938
|
Babam1
|
BRISC and BRCA1 A complex member 1 |
| chr8_-_122987191 | 0.02 |
ENSRNOT00000033976
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr10_-_70681672 | 0.02 |
ENSRNOT00000013836
|
Mmp28
|
matrix metallopeptidase 28 |
| chr4_+_56337695 | 0.02 |
ENSRNOT00000071926
|
Lep
|
leptin |
| chr7_-_126691232 | 0.02 |
ENSRNOT00000055909
|
Cdpf1
|
cysteine rich, DPF motif domain containing 1 |
| chr3_+_72238981 | 0.02 |
ENSRNOT00000011006
|
Slc43a1
|
solute carrier family 43 member 1 |
| chr8_-_47339343 | 0.02 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr15_-_38276159 | 0.02 |
ENSRNOT00000084698
ENSRNOT00000015233 |
Micu2
|
mitochondrial calcium uptake 2 |
| chr2_+_248219428 | 0.02 |
ENSRNOT00000037181
|
LOC685067
|
similar to guanylate binding protein family, member 6 |
| chr2_+_188516582 | 0.02 |
ENSRNOT00000074727
|
Gba
|
glucosylceramidase beta |
| chr3_-_14395362 | 0.02 |
ENSRNOT00000092787
|
Rab14
|
RAB14, member RAS oncogene family |
| chr14_+_83298674 | 0.02 |
ENSRNOT00000024813
|
Pisd
|
phosphatidylserine decarboxylase |
| chr8_+_26432514 | 0.02 |
ENSRNOT00000081574
|
Sept7
|
septin 7 |
| chr10_+_106712127 | 0.02 |
ENSRNOT00000040629
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr15_-_8989580 | 0.02 |
ENSRNOT00000061402
|
Thrb
|
thyroid hormone receptor beta |
| chr18_+_30904498 | 0.02 |
ENSRNOT00000026969
|
Pcdhga11
|
protocadherin gamma subfamily A, 11 |
| chr4_-_79015660 | 0.02 |
ENSRNOT00000068441
|
LOC100910678
|
coiled-coil domain-containing protein 72-like |
| chr20_-_45024315 | 0.02 |
ENSRNOT00000066856
|
RGD1304770
|
similar to Na+ dependent glucose transporter 1 |
| chr5_+_48561339 | 0.02 |
ENSRNOT00000010614
ENSRNOT00000091827 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr7_+_141702038 | 0.02 |
ENSRNOT00000086577
|
Dip2b
|
disco-interacting protein 2 homolog B |
| chr4_-_7885301 | 0.02 |
ENSRNOT00000035034
|
Rint1
|
RAD50 interactor 1 |
| chr5_+_25349928 | 0.02 |
ENSRNOT00000020826
|
Gem
|
GTP binding protein overexpressed in skeletal muscle |
| chrX_+_20216587 | 0.02 |
ENSRNOT00000073114
|
AABR07037412.2
|
FYVE, RhoGEF and PH domain-containing protein 1 |
| chr9_-_85243001 | 0.02 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr12_+_48316143 | 0.02 |
ENSRNOT00000084511
|
Svop
|
SV2 related protein |
| chr13_-_95250235 | 0.02 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr15_-_4454958 | 0.01 |
ENSRNOT00000070933
|
Nudt13
|
nudix hydrolase 13 |
| chr18_+_44468784 | 0.01 |
ENSRNOT00000031812
|
Dmxl1
|
Dmx-like 1 |
| chr8_+_116054465 | 0.01 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr18_+_69841053 | 0.01 |
ENSRNOT00000071545
ENSRNOT00000030613 ENSRNOT00000075543 |
Mro
|
maestro |
| chr4_+_2505909 | 0.01 |
ENSRNOT00000092756
|
Ube3c
|
ubiquitin protein ligase E3C |
| chr3_-_162581610 | 0.01 |
ENSRNOT00000079324
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr8_+_49282460 | 0.01 |
ENSRNOT00000021488
|
Cd3d
|
CD3d molecule |
| chr13_-_88307988 | 0.01 |
ENSRNOT00000003812
|
Hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chrX_-_135250519 | 0.01 |
ENSRNOT00000044487
|
Elf4
|
E74 like ETS transcription factor 4 |
| chr3_-_81367630 | 0.01 |
ENSRNOT00000067509
|
Slc35c1
|
solute carrier family 35 member C1 |
| chr4_+_22084954 | 0.01 |
ENSRNOT00000090968
|
Crot
|
carnitine O-octanoyltransferase |
| chr1_-_225952516 | 0.01 |
ENSRNOT00000043387
|
Incenp
|
inner centromere protein |
| chr20_-_821332 | 0.01 |
ENSRNOT00000040957
|
Olr1692
|
olfactory receptor 1692 |
| chr9_+_97355924 | 0.01 |
ENSRNOT00000026558
|
Ackr3
|
atypical chemokine receptor 3 |
| chr17_+_9797907 | 0.01 |
ENSRNOT00000021638
|
Lman2
|
lectin, mannose-binding 2 |
| chr6_-_114488880 | 0.01 |
ENSRNOT00000087560
|
AC118957.1
|
|
| chr19_+_55197704 | 0.01 |
ENSRNOT00000045657
|
Zc3h18
|
zinc finger CCCH-type containing 18 |
| chr3_-_171893901 | 0.01 |
ENSRNOT00000091966
|
Apcdd1l
|
APC down-regulated 1 like |
| chr6_+_28235695 | 0.01 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr8_+_129201669 | 0.01 |
ENSRNOT00000025663
|
Entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr10_+_39435227 | 0.01 |
ENSRNOT00000042144
ENSRNOT00000077185 |
P4ha2
|
prolyl 4-hydroxylase subunit alpha 2 |
| chr8_-_116764780 | 0.01 |
ENSRNOT00000026216
|
Fam212a
|
family with sequence similarity 212, member A |
| chr18_+_63882610 | 0.01 |
ENSRNOT00000022717
|
Ldlrad4
|
low density lipoprotein receptor class A domain containing 4 |
| chr17_-_14679409 | 0.01 |
ENSRNOT00000020704
|
Aspn
|
asporin |
| chrX_-_138435391 | 0.01 |
ENSRNOT00000043258
|
Mbnl3
|
muscleblind-like splicing regulator 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Meis1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.1 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.0 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.0 | 0.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:2001013 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.0 | GO:1990478 | response to ultrasound(GO:1990478) |
| 0.0 | 0.2 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.0 | GO:1990907 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.0 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |