Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
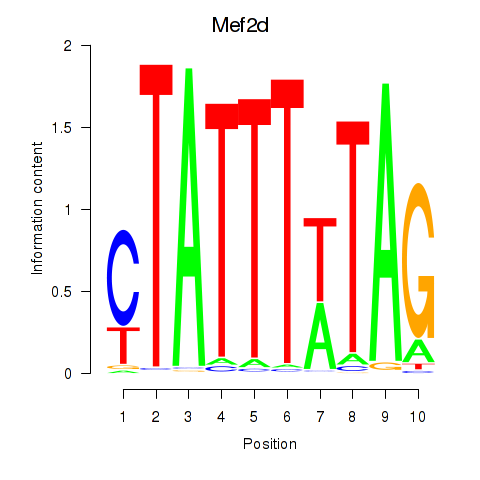
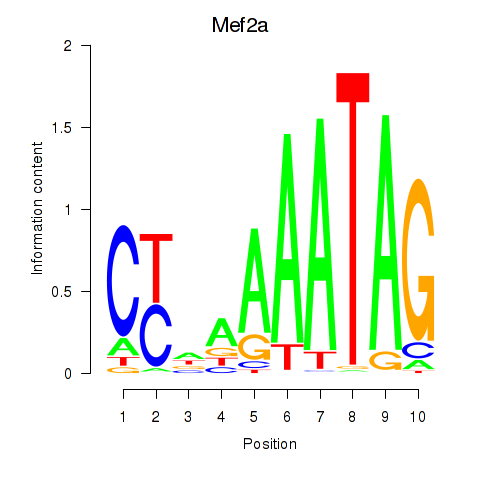
Results for Mef2d_Mef2a
Z-value: 0.42
Transcription factors associated with Mef2d_Mef2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2d
|
ENSRNOG00000031778 | myocyte enhancer factor 2D |
|
Mef2a
|
ENSRNOG00000047756 | myocyte enhancer factor 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2d | rn6_v1_chr2_+_187512164_187512164 | 0.61 | 2.7e-01 | Click! |
| Mef2a | rn6_v1_chr1_-_128287151_128287151 | 0.44 | 4.6e-01 | Click! |
Activity profile of Mef2d_Mef2a motif
Sorted Z-values of Mef2d_Mef2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_73827488 | 0.35 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr4_+_148782479 | 0.33 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr18_-_26211445 | 0.32 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr2_+_196334626 | 0.30 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chrX_-_40086870 | 0.23 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr3_-_170040953 | 0.19 |
ENSRNOT00000005866
|
Cbln4
|
cerebellin 4 precursor |
| chr16_+_72388880 | 0.19 |
ENSRNOT00000072459
|
LOC684871
|
similar to Protein C8orf4 (Thyroid cancer protein 1) (TC-1) |
| chr2_+_204512302 | 0.18 |
ENSRNOT00000021846
|
Casq2
|
calsequestrin 2 |
| chr1_-_25839198 | 0.18 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr2_-_45518502 | 0.15 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr12_-_23841049 | 0.14 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr10_-_56558487 | 0.13 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr5_-_114014277 | 0.11 |
ENSRNOT00000011731
|
Jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr3_+_150323116 | 0.11 |
ENSRNOT00000023429
ENSRNOT00000080485 |
Raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr10_-_88670430 | 0.11 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr1_-_199624783 | 0.10 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chrX_+_71528988 | 0.10 |
ENSRNOT00000075963
|
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr1_+_101178104 | 0.10 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr1_+_199449973 | 0.08 |
ENSRNOT00000029994
|
Trim72
|
tripartite motif containing 72 |
| chr4_+_153874852 | 0.08 |
ENSRNOT00000079744
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr10_-_32471454 | 0.08 |
ENSRNOT00000003224
|
Sgcd
|
sarcoglycan, delta |
| chr1_-_264975132 | 0.08 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr13_-_90710148 | 0.08 |
ENSRNOT00000010113
ENSRNOT00000080638 |
Kcnj9
|
potassium voltage-gated channel subfamily J member 9 |
| chr7_+_142912316 | 0.08 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_+_238222521 | 0.07 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chrX_+_33895769 | 0.07 |
ENSRNOT00000085993
ENSRNOT00000079365 |
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr1_+_31531143 | 0.07 |
ENSRNOT00000034704
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr10_+_55687050 | 0.07 |
ENSRNOT00000057136
|
Per1
|
period circadian clock 1 |
| chr3_+_58965552 | 0.07 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr11_-_34526931 | 0.07 |
ENSRNOT00000002291
|
Hlcs
|
holocarboxylase synthetase |
| chr8_+_110982777 | 0.07 |
ENSRNOT00000010992
|
Ky
|
kyphoscoliosis peptidase |
| chr5_-_22769907 | 0.06 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr10_+_63803309 | 0.06 |
ENSRNOT00000036666
|
Myo1c
|
myosin 1C |
| chr5_+_147257678 | 0.06 |
ENSRNOT00000066268
|
Rnf19b
|
ring finger protein 19B |
| chr6_+_73553210 | 0.06 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr10_-_4257868 | 0.06 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr5_+_152533349 | 0.06 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr8_+_117836701 | 0.05 |
ENSRNOT00000043345
|
Plxnb1
|
plexin B1 |
| chr3_+_161433410 | 0.05 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chr2_-_172361779 | 0.05 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr4_-_81241152 | 0.05 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr10_-_16046033 | 0.05 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_-_247988462 | 0.05 |
ENSRNOT00000022387
|
Pdlim5
|
PDZ and LIM domain 5 |
| chr9_-_9702306 | 0.05 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr2_-_227411964 | 0.05 |
ENSRNOT00000019931
|
Synpo2
|
synaptopodin 2 |
| chr4_-_179512471 | 0.04 |
ENSRNOT00000012588
|
Kras
|
KRAS proto-oncogene, GTPase |
| chr3_+_151126591 | 0.04 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr3_+_79940561 | 0.04 |
ENSRNOT00000016652
|
Mybpc3
|
myosin binding protein C, cardiac |
| chr18_+_63203063 | 0.04 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr15_+_4064706 | 0.04 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr13_-_90602365 | 0.04 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr13_-_51076165 | 0.04 |
ENSRNOT00000004602
|
Adora1
|
adenosine A1 receptor |
| chr18_+_32273770 | 0.04 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr7_+_142905758 | 0.04 |
ENSRNOT00000078663
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_-_16045835 | 0.03 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr13_-_99531959 | 0.03 |
ENSRNOT00000005059
|
Wdr26
|
WD repeat domain 26 |
| chr3_+_56868167 | 0.03 |
ENSRNOT00000087134
|
Gad1
|
glutamate decarboxylase 1 |
| chr3_+_173955686 | 0.03 |
ENSRNOT00000088447
|
Fam217b
|
family with sequence similarity 217, member B |
| chr9_+_82571269 | 0.03 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr2_+_219598162 | 0.03 |
ENSRNOT00000020297
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr5_-_144274981 | 0.03 |
ENSRNOT00000065871
|
Map7d1
|
MAP7 domain containing 1 |
| chr5_+_151776004 | 0.03 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr8_-_55177818 | 0.03 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr1_+_100299626 | 0.03 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr16_-_74264142 | 0.03 |
ENSRNOT00000026067
|
Dkk4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr2_-_104958034 | 0.03 |
ENSRNOT00000080699
|
Gyg1
|
glycogenin 1 |
| chr9_+_82556573 | 0.03 |
ENSRNOT00000026860
|
Des
|
desmin |
| chr2_-_33025271 | 0.03 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_111818150 | 0.02 |
ENSRNOT00000067181
|
Jmjd7
|
jumonji domain containing 7 |
| chr1_-_142164007 | 0.02 |
ENSRNOT00000078982
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr3_-_5575136 | 0.02 |
ENSRNOT00000008034
|
Slc2a6
|
solute carrier family 2 member 6 |
| chr10_-_65502936 | 0.02 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr7_+_35309265 | 0.02 |
ENSRNOT00000080291
|
Tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr10_-_86688730 | 0.02 |
ENSRNOT00000055333
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr2_+_93792601 | 0.02 |
ENSRNOT00000014701
ENSRNOT00000077311 |
Fabp4
|
fatty acid binding protein 4 |
| chr8_+_130416355 | 0.02 |
ENSRNOT00000026234
|
Klhl40
|
kelch-like family member 40 |
| chr12_+_17734133 | 0.02 |
ENSRNOT00000042117
|
Pdgfa
|
platelet derived growth factor subunit A |
| chr17_+_57075218 | 0.02 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr2_+_198772937 | 0.02 |
ENSRNOT00000028812
|
Itga10
|
integrin subunit alpha 10 |
| chr13_+_52624878 | 0.02 |
ENSRNOT00000076054
ENSRNOT00000076299 |
Tnni1
|
troponin I1, slow skeletal type |
| chr4_-_100303047 | 0.02 |
ENSRNOT00000018170
ENSRNOT00000084782 |
Mat2a
|
methionine adenosyltransferase 2A |
| chr1_-_166912524 | 0.02 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr16_+_49266903 | 0.02 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr2_-_187854363 | 0.02 |
ENSRNOT00000092993
|
Lmna
|
lamin A/C |
| chr2_+_212257225 | 0.02 |
ENSRNOT00000077883
|
Vav3
|
vav guanine nucleotide exchange factor 3 |
| chr10_+_90217924 | 0.02 |
ENSRNOT00000028388
|
Asb16
|
ankyrin repeat and SOCS box-containing 16 |
| chr1_-_56683731 | 0.02 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr15_-_28314459 | 0.02 |
ENSRNOT00000042055
ENSRNOT00000040540 |
Ndrg2
|
NDRG family member 2 |
| chr9_-_97151832 | 0.02 |
ENSRNOT00000040169
|
Asb18
|
ankyrin repeat and SOCS box-containing 18 |
| chr4_+_71675383 | 0.02 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr14_+_63095720 | 0.02 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr5_-_150704117 | 0.02 |
ENSRNOT00000067455
|
Sesn2
|
sestrin 2 |
| chr9_-_113504605 | 0.02 |
ENSRNOT00000067974
|
Rab31
|
RAB31, member RAS oncogene family |
| chr6_-_136145837 | 0.02 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr1_-_142164263 | 0.02 |
ENSRNOT00000016281
|
Man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr13_+_99136871 | 0.01 |
ENSRNOT00000078263
ENSRNOT00000004350 |
Sde2
|
SDE2 telomere maintenance homolog |
| chr13_-_81214494 | 0.01 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr10_+_89459650 | 0.01 |
ENSRNOT00000028149
ENSRNOT00000077942 |
Nbr1
|
NBR1, autophagy cargo receptor |
| chr10_+_3218466 | 0.01 |
ENSRNOT00000093629
ENSRNOT00000093338 ENSRNOT00000077695 |
Ntan1
|
N-terminal asparagine amidase |
| chr11_-_90406797 | 0.01 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr17_-_43689311 | 0.01 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chr17_+_72218769 | 0.01 |
ENSRNOT00000041346
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr3_+_65816569 | 0.01 |
ENSRNOT00000079672
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr10_-_70871066 | 0.01 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr1_-_282170017 | 0.01 |
ENSRNOT00000066947
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr2_-_198016898 | 0.01 |
ENSRNOT00000025523
|
Car14
|
carbonic anhydrase 14 |
| chr3_+_5709236 | 0.01 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr10_+_86337728 | 0.01 |
ENSRNOT00000085408
|
Tcap
|
titin-cap |
| chr2_-_197991198 | 0.01 |
ENSRNOT00000056322
|
Ciart
|
circadian associated repressor of transcription |
| chr2_-_157759819 | 0.01 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr3_+_79918969 | 0.01 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr7_-_123767797 | 0.01 |
ENSRNOT00000012699
|
Tcf20
|
transcription factor 20 |
| chr1_-_199270627 | 0.01 |
ENSRNOT00000026063
|
Stx1b
|
syntaxin 1B |
| chr2_-_178057157 | 0.01 |
ENSRNOT00000091135
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr2_-_197991574 | 0.01 |
ENSRNOT00000085632
|
Ciart
|
circadian associated repressor of transcription |
| chr3_+_122114108 | 0.01 |
ENSRNOT00000091935
|
Sirpa
|
signal-regulatory protein alpha |
| chr17_-_43614844 | 0.01 |
ENSRNOT00000023054
|
Hist1h1a
|
histone cluster 1 H1 family member a |
| chr20_+_13940877 | 0.01 |
ENSRNOT00000093587
|
Cabin1
|
calcineurin binding protein 1 |
| chr2_+_198721724 | 0.01 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr4_+_83391283 | 0.01 |
ENSRNOT00000031365
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr7_-_132143470 | 0.01 |
ENSRNOT00000038946
ENSRNOT00000044092 ENSRNOT00000066528 ENSRNOT00000045553 ENSRNOT00000046744 ENSRNOT00000055742 |
Kif21a
|
kinesin family member 21A |
| chr2_+_184230459 | 0.01 |
ENSRNOT00000074187
|
AABR07012054.1
|
|
| chr13_+_52662996 | 0.01 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr9_-_52238564 | 0.01 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr15_+_3996048 | 0.00 |
ENSRNOT00000078653
|
Ndst2
|
N-deacetylase and N-sulfotransferase 2 |
| chr9_+_47536824 | 0.00 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr10_+_53740841 | 0.00 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr4_-_50312608 | 0.00 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr1_+_21748261 | 0.00 |
ENSRNOT00000019519
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr5_+_173640780 | 0.00 |
ENSRNOT00000027476
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr13_-_77896697 | 0.00 |
ENSRNOT00000003452
|
Tnn
|
tenascin N |
| chr14_-_17171580 | 0.00 |
ENSRNOT00000085525
|
Art3
|
ADP-ribosyltransferase 3 |
| chrX_-_138972684 | 0.00 |
ENSRNOT00000040165
|
Hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr16_+_2537248 | 0.00 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr1_+_265904566 | 0.00 |
ENSRNOT00000086041
ENSRNOT00000036156 |
Gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr5_+_154598758 | 0.00 |
ENSRNOT00000015776
|
Tcea3
|
transcription elongation factor A3 |
| chr10_+_82032656 | 0.00 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr20_-_20713128 | 0.00 |
ENSRNOT00000000784
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr7_+_144628120 | 0.00 |
ENSRNOT00000022247
|
Hoxc5
|
homeo box C5 |
| chr10_-_82785142 | 0.00 |
ENSRNOT00000005381
|
Sgca
|
sarcoglycan, alpha |
| chr8_-_127782070 | 0.00 |
ENSRNOT00000045493
|
Plcd1
|
phospholipase C, delta 1 |
| chr15_-_33250546 | 0.00 |
ENSRNOT00000017857
|
RGD1565222
|
similar to RIKEN cDNA 4931414P19 |
| chr5_-_152589719 | 0.00 |
ENSRNOT00000022522
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr1_-_89488223 | 0.00 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr7_-_14402010 | 0.00 |
ENSRNOT00000008303
|
Wiz
|
widely-interspaced zinc finger motifs |
| chr7_-_107391184 | 0.00 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr14_+_83897117 | 0.00 |
ENSRNOT00000026569
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr20_-_31597830 | 0.00 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2d_Mef2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:1903544 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 0.0 | 0.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:0070256 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 0.0 | 0.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.0 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |