Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
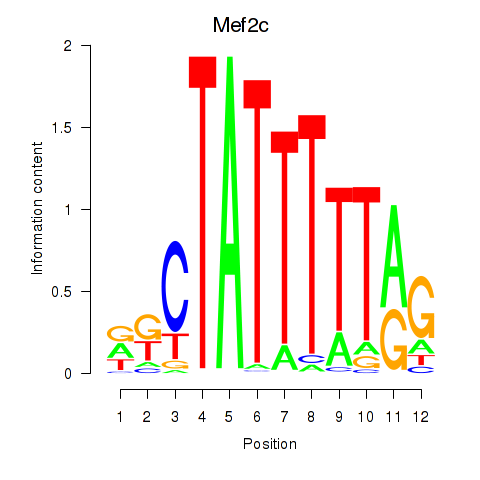
Results for Mef2c
Z-value: 0.34
Transcription factors associated with Mef2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2c
|
ENSRNOG00000033134 | myocyte enhancer factor 2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2c | rn6_v1_chr2_+_11658568_11658568 | -0.06 | 9.2e-01 | Click! |
Activity profile of Mef2c motif
Sorted Z-values of Mef2c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_142912316 | 0.24 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_25839198 | 0.19 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr20_-_22459025 | 0.14 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr16_-_73827488 | 0.13 |
ENSRNOT00000064070
|
Ank1
|
ankyrin 1 |
| chr3_-_170040953 | 0.12 |
ENSRNOT00000005866
|
Cbln4
|
cerebellin 4 precursor |
| chr1_+_215666628 | 0.10 |
ENSRNOT00000040598
ENSRNOT00000066135 ENSRNOT00000051425 ENSRNOT00000080339 ENSRNOT00000066896 ENSRNOT00000063918 |
Tnnt3
|
troponin T3, fast skeletal type |
| chr2_-_45518502 | 0.10 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr18_-_26211445 | 0.10 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr1_+_219250265 | 0.10 |
ENSRNOT00000024353
|
Doc2g
|
double C2-like domains, gamma |
| chr10_-_76039964 | 0.09 |
ENSRNOT00000003164
|
Msi2
|
musashi RNA-binding protein 2 |
| chr1_-_199624783 | 0.08 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr12_-_23841049 | 0.08 |
ENSRNOT00000031555
|
Hspb1
|
heat shock protein family B (small) member 1 |
| chr13_+_52624878 | 0.08 |
ENSRNOT00000076054
ENSRNOT00000076299 |
Tnni1
|
troponin I1, slow skeletal type |
| chr3_+_58965552 | 0.08 |
ENSRNOT00000002068
|
Map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr1_+_31531143 | 0.07 |
ENSRNOT00000034704
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr5_-_114014277 | 0.07 |
ENSRNOT00000011731
|
Jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr2_+_204512302 | 0.07 |
ENSRNOT00000021846
|
Casq2
|
calsequestrin 2 |
| chr11_+_9642365 | 0.06 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr10_-_4257868 | 0.06 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr1_-_166912524 | 0.06 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr5_+_147257678 | 0.06 |
ENSRNOT00000066268
|
Rnf19b
|
ring finger protein 19B |
| chr10_-_86688730 | 0.06 |
ENSRNOT00000055333
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr1_+_100299626 | 0.05 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chrX_+_33895769 | 0.05 |
ENSRNOT00000085993
ENSRNOT00000079365 |
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr3_+_151126591 | 0.05 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr16_+_72388880 | 0.05 |
ENSRNOT00000072459
|
LOC684871
|
similar to Protein C8orf4 (Thyroid cancer protein 1) (TC-1) |
| chr1_+_21748261 | 0.05 |
ENSRNOT00000019519
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr2_+_198772937 | 0.05 |
ENSRNOT00000028812
|
Itga10
|
integrin subunit alpha 10 |
| chr1_+_101178104 | 0.05 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chr5_+_151776004 | 0.05 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr11_+_45888221 | 0.04 |
ENSRNOT00000071932
ENSRNOT00000066008 ENSRNOT00000060802 |
Lnp1
|
leukemia NUP98 fusion partner 1 |
| chr13_-_90710148 | 0.04 |
ENSRNOT00000010113
ENSRNOT00000080638 |
Kcnj9
|
potassium voltage-gated channel subfamily J member 9 |
| chr11_-_90406797 | 0.04 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr6_+_73553210 | 0.04 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr3_+_79918969 | 0.04 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr2_+_154604832 | 0.04 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr5_+_154598758 | 0.04 |
ENSRNOT00000015776
|
Tcea3
|
transcription elongation factor A3 |
| chr15_+_34187223 | 0.04 |
ENSRNOT00000024978
|
Cpne6
|
copine 6 |
| chr17_-_43422846 | 0.04 |
ENSRNOT00000050526
|
Hist1h2aa
|
histone cluster 1 H2A family member a |
| chr3_+_56868167 | 0.04 |
ENSRNOT00000087134
|
Gad1
|
glutamate decarboxylase 1 |
| chr3_+_161433410 | 0.04 |
ENSRNOT00000024657
|
Slc12a5
|
solute carrier family 12 member 5 |
| chrX_+_71528988 | 0.04 |
ENSRNOT00000075963
|
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chrX_+_159158194 | 0.03 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr13_+_52662996 | 0.03 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr9_+_111028824 | 0.03 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr13_-_81214494 | 0.03 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr2_+_242634399 | 0.03 |
ENSRNOT00000035700
|
Emcn
|
endomucin |
| chr3_-_123119460 | 0.03 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chr10_+_86337728 | 0.03 |
ENSRNOT00000085408
|
Tcap
|
titin-cap |
| chr2_-_172361779 | 0.03 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr17_+_57075218 | 0.03 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr10_+_54352270 | 0.03 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr3_+_110442637 | 0.03 |
ENSRNOT00000010471
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr4_+_153874852 | 0.03 |
ENSRNOT00000079744
|
Slc6a13
|
solute carrier family 6 member 13 |
| chr6_-_76608864 | 0.03 |
ENSRNOT00000010824
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr11_-_74315248 | 0.03 |
ENSRNOT00000002346
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr2_-_247988462 | 0.03 |
ENSRNOT00000022387
|
Pdlim5
|
PDZ and LIM domain 5 |
| chr5_-_160219812 | 0.03 |
ENSRNOT00000016333
|
Plekhm2
|
pleckstrin homology and RUN domain containing M2 |
| chr17_-_55346279 | 0.02 |
ENSRNOT00000025037
|
Svil
|
supervillin |
| chr9_-_9702306 | 0.02 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr13_+_99136871 | 0.02 |
ENSRNOT00000078263
ENSRNOT00000004350 |
Sde2
|
SDE2 telomere maintenance homolog |
| chr5_-_22769907 | 0.02 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr2_-_104958034 | 0.02 |
ENSRNOT00000080699
|
Gyg1
|
glycogenin 1 |
| chr5_-_139748489 | 0.02 |
ENSRNOT00000078741
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr11_+_86520992 | 0.02 |
ENSRNOT00000040954
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr2_+_198721724 | 0.02 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr7_-_114305896 | 0.02 |
ENSRNOT00000067327
ENSRNOT00000038172 |
Trappc9
|
trafficking protein particle complex 9 |
| chr3_+_150323116 | 0.02 |
ENSRNOT00000023429
ENSRNOT00000080485 |
Raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr1_-_56683731 | 0.02 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr3_-_36660758 | 0.02 |
ENSRNOT00000006111
|
Rnd3
|
Rho family GTPase 3 |
| chr7_+_70807867 | 0.02 |
ENSRNOT00000010639
|
Stac3
|
SH3 and cysteine rich domain 3 |
| chr1_-_128341240 | 0.02 |
ENSRNOT00000070864
ENSRNOT00000072915 |
Mef2a
|
myocyte enhancer factor 2a |
| chr5_+_25042710 | 0.02 |
ENSRNOT00000061385
|
RGD1559904
|
similar to mKIAA1429 protein |
| chr2_+_238257031 | 0.02 |
ENSRNOT00000016066
|
Ints12
|
integrator complex subunit 12 |
| chr9_-_81864202 | 0.02 |
ENSRNOT00000084964
|
Zfp142
|
zinc finger protein 142 |
| chr10_+_90217924 | 0.02 |
ENSRNOT00000028388
|
Asb16
|
ankyrin repeat and SOCS box-containing 16 |
| chr15_-_33250546 | 0.02 |
ENSRNOT00000017857
|
RGD1565222
|
similar to RIKEN cDNA 4931414P19 |
| chr3_+_122114108 | 0.02 |
ENSRNOT00000091935
|
Sirpa
|
signal-regulatory protein alpha |
| chr2_-_178057157 | 0.02 |
ENSRNOT00000091135
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr14_-_17171580 | 0.02 |
ENSRNOT00000085525
|
Art3
|
ADP-ribosyltransferase 3 |
| chr12_+_17734133 | 0.01 |
ENSRNOT00000042117
|
Pdgfa
|
platelet derived growth factor subunit A |
| chr13_-_99531959 | 0.01 |
ENSRNOT00000005059
|
Wdr26
|
WD repeat domain 26 |
| chr15_+_39779648 | 0.01 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr5_+_148528725 | 0.01 |
ENSRNOT00000017325
|
Fabp3
|
fatty acid binding protein 3 |
| chr9_+_82571269 | 0.01 |
ENSRNOT00000026941
|
Speg
|
SPEG complex locus |
| chr8_+_130416355 | 0.01 |
ENSRNOT00000026234
|
Klhl40
|
kelch-like family member 40 |
| chr2_+_32820322 | 0.01 |
ENSRNOT00000013768
|
Cd180
|
CD180 molecule |
| chr8_-_76579099 | 0.01 |
ENSRNOT00000088628
|
Fam81a
|
family with sequence similarity 81, member A |
| chr4_-_161850875 | 0.01 |
ENSRNOT00000009467
|
Pzp
|
pregnancy-zone protein |
| chr10_+_89459650 | 0.01 |
ENSRNOT00000028149
ENSRNOT00000077942 |
Nbr1
|
NBR1, autophagy cargo receptor |
| chr15_-_33428942 | 0.01 |
ENSRNOT00000019618
ENSRNOT00000092160 |
Slc7a8
|
solute carrier family 7 member 8 |
| chr10_-_16046033 | 0.01 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chrX_-_9999401 | 0.01 |
ENSRNOT00000060992
|
Gpr82
|
G protein-coupled receptor 82 |
| chr2_+_219598162 | 0.01 |
ENSRNOT00000020297
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr10_-_56558487 | 0.01 |
ENSRNOT00000023256
|
Slc2a4
|
solute carrier family 2 member 4 |
| chr10_+_82032656 | 0.01 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr10_+_3218466 | 0.01 |
ENSRNOT00000093629
ENSRNOT00000093338 ENSRNOT00000077695 |
Ntan1
|
N-terminal asparagine amidase |
| chr5_-_152589719 | 0.01 |
ENSRNOT00000022522
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr9_+_47536824 | 0.01 |
ENSRNOT00000049349
|
Tmem182
|
transmembrane protein 182 |
| chr18_+_63203063 | 0.01 |
ENSRNOT00000024144
|
Prelid3a
|
PRELI domain containing 3A |
| chr1_-_221370322 | 0.01 |
ENSRNOT00000028431
|
Capn1
|
calpain 1 |
| chr8_-_30222036 | 0.01 |
ENSRNOT00000035454
|
Ntm
|
neurotrimin |
| chr7_+_11383116 | 0.01 |
ENSRNOT00000066348
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr14_-_43072843 | 0.01 |
ENSRNOT00000064263
|
Limch1
|
LIM and calponin homology domains 1 |
| chr3_+_5709236 | 0.01 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr10_-_62699723 | 0.01 |
ENSRNOT00000086706
|
Coro6
|
coronin 6 |
| chr5_+_58995249 | 0.01 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr8_+_67295727 | 0.01 |
ENSRNOT00000020443
|
Anp32a
|
acidic nuclear phosphoprotein 32 family member A |
| chr7_-_107391184 | 0.01 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr18_+_73256085 | 0.01 |
ENSRNOT00000080660
|
Hdhd2
|
haloacid dehalogenase-like hydrolase domain containing 2 |
| chr17_+_36334147 | 0.01 |
ENSRNOT00000050261
|
E2f3
|
E2F transcription factor 3 |
| chr8_+_110982777 | 0.00 |
ENSRNOT00000010992
|
Ky
|
kyphoscoliosis peptidase |
| chr9_-_113504605 | 0.00 |
ENSRNOT00000067974
|
Rab31
|
RAB31, member RAS oncogene family |
| chr2_-_261337163 | 0.00 |
ENSRNOT00000030341
|
Tnni3k
|
TNNI3 interacting kinase |
| chr5_+_152533349 | 0.00 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr7_+_80351774 | 0.00 |
ENSRNOT00000081948
|
Oxr1
|
oxidation resistance 1 |
| chr13_-_90602365 | 0.00 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr7_+_35309265 | 0.00 |
ENSRNOT00000080291
|
Tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr9_-_81772851 | 0.00 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr3_+_79940561 | 0.00 |
ENSRNOT00000016652
|
Mybpc3
|
myosin binding protein C, cardiac |
| chr4_+_14001761 | 0.00 |
ENSRNOT00000076519
|
Cd36
|
CD36 molecule |
| chr10_-_16045835 | 0.00 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.0 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |