Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
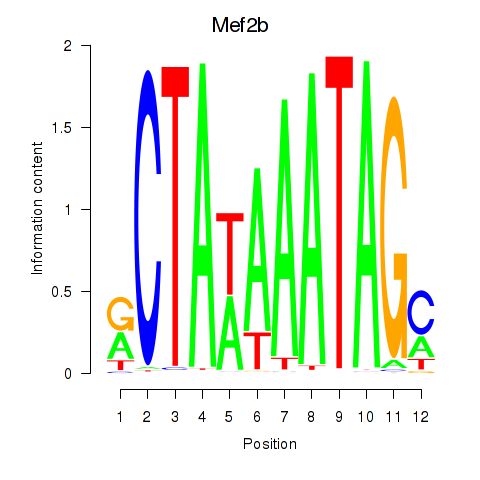
Results for Mef2b
Z-value: 0.33
Transcription factors associated with Mef2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2b
|
ENSRNOG00000020400 | myocyte enhancer factor 2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2b | rn6_v1_chr16_-_21017163_21017163 | 0.48 | 4.1e-01 | Click! |
Activity profile of Mef2b motif
Sorted Z-values of Mef2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_15274917 | 0.39 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr7_+_142912316 | 0.27 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_+_101178104 | 0.14 |
ENSRNOT00000028072
|
Pth2
|
parathyroid hormone 2 |
| chrX_+_13117239 | 0.13 |
ENSRNOT00000088998
|
AABR07037112.1
|
|
| chr3_-_170040953 | 0.12 |
ENSRNOT00000005866
|
Cbln4
|
cerebellin 4 precursor |
| chr2_-_45518502 | 0.11 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chrX_-_40086870 | 0.09 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr3_+_147358858 | 0.08 |
ENSRNOT00000012951
|
Rspo4
|
R-spondin 4 |
| chr2_+_196334626 | 0.08 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr1_+_31531143 | 0.08 |
ENSRNOT00000034704
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chrX_-_13116743 | 0.08 |
ENSRNOT00000004305
|
Mid1ip1
|
MID1 interacting protein 1 |
| chr13_+_53283855 | 0.08 |
ENSRNOT00000043590
|
RGD1564839
|
similar to ribosomal protein L31 |
| chr18_+_55505993 | 0.07 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr17_+_57075218 | 0.07 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr18_+_55463308 | 0.06 |
ENSRNOT00000073388
|
LOC100910979
|
interferon-inducible GTPase 1-like |
| chr1_-_199624783 | 0.06 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr11_-_1983513 | 0.06 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chr3_+_56868167 | 0.06 |
ENSRNOT00000087134
|
Gad1
|
glutamate decarboxylase 1 |
| chr2_-_53185926 | 0.06 |
ENSRNOT00000081558
|
Ghr
|
growth hormone receptor |
| chr17_-_389967 | 0.06 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr10_-_70871066 | 0.06 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr11_+_46184871 | 0.06 |
ENSRNOT00000048417
|
Tfg
|
Trk-fused gene |
| chr5_+_151776004 | 0.06 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr17_-_43807540 | 0.06 |
ENSRNOT00000074763
|
LOC684762
|
similar to CG31613-PA |
| chr1_-_47307488 | 0.06 |
ENSRNOT00000090033
|
Ezr
|
ezrin |
| chr10_+_54352270 | 0.06 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr10_+_90217924 | 0.06 |
ENSRNOT00000028388
|
Asb16
|
ankyrin repeat and SOCS box-containing 16 |
| chr1_-_153752541 | 0.05 |
ENSRNOT00000080927
|
Prss23
|
protease, serine, 23 |
| chr5_-_114014277 | 0.05 |
ENSRNOT00000011731
|
Jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr9_+_98490608 | 0.05 |
ENSRNOT00000027232
|
Klhl30
|
kelch-like family member 30 |
| chr5_-_152473868 | 0.05 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr18_-_26211445 | 0.05 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr4_+_7258176 | 0.05 |
ENSRNOT00000061992
|
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr10_+_3218466 | 0.05 |
ENSRNOT00000093629
ENSRNOT00000093338 ENSRNOT00000077695 |
Ntan1
|
N-terminal asparagine amidase |
| chr14_-_17171580 | 0.05 |
ENSRNOT00000085525
|
Art3
|
ADP-ribosyltransferase 3 |
| chr14_-_19072677 | 0.04 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr5_+_145188323 | 0.04 |
ENSRNOT00000038888
|
Tmem35b
|
transmembrane protein 35B |
| chr1_-_20155960 | 0.04 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr6_-_60124274 | 0.04 |
ENSRNOT00000059823
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr17_+_76002275 | 0.04 |
ENSRNOT00000092665
ENSRNOT00000086701 |
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr2_+_198772937 | 0.04 |
ENSRNOT00000028812
|
Itga10
|
integrin subunit alpha 10 |
| chr1_+_80279706 | 0.04 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr19_+_56272162 | 0.04 |
ENSRNOT00000030399
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (S. cerevisiae) |
| chr5_-_22769907 | 0.04 |
ENSRNOT00000047805
ENSRNOT00000076167 ENSRNOT00000076507 ENSRNOT00000076113 ENSRNOT00000083779 |
Asph
|
aspartate-beta-hydroxylase |
| chr1_+_55219773 | 0.04 |
ENSRNOT00000041610
|
RGD1561667
|
similar to putative protein kinase |
| chr16_+_54386513 | 0.04 |
ENSRNOT00000080696
|
AABR07025896.1
|
|
| chr3_+_79918969 | 0.04 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr11_-_34526931 | 0.04 |
ENSRNOT00000002291
|
Hlcs
|
holocarboxylase synthetase |
| chr4_+_153874852 | 0.04 |
ENSRNOT00000079744
|
Slc6a13
|
solute carrier family 6 member 13 |
| chrM_-_14061 | 0.04 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_+_169590308 | 0.04 |
ENSRNOT00000023147
|
Olr155
|
olfactory receptor 155 |
| chr2_+_185846232 | 0.03 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr19_+_10731855 | 0.03 |
ENSRNOT00000022277
|
Pllp
|
plasmolipin |
| chr2_-_172361779 | 0.03 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr4_+_88328061 | 0.03 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr5_+_147257678 | 0.03 |
ENSRNOT00000066268
|
Rnf19b
|
ring finger protein 19B |
| chr4_-_50312608 | 0.03 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr10_-_43512287 | 0.03 |
ENSRNOT00000036481
|
Faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr13_-_81214494 | 0.03 |
ENSRNOT00000004950
ENSRNOT00000082385 |
Prrx1
|
paired related homeobox 1 |
| chr4_+_119680112 | 0.03 |
ENSRNOT00000013282
|
Gp9
|
glycoprotein IX (platelet) |
| chr17_-_43422846 | 0.03 |
ENSRNOT00000050526
|
Hist1h2aa
|
histone cluster 1 H2A family member a |
| chr4_-_49422824 | 0.03 |
ENSRNOT00000077889
|
Fam3c
|
family with sequence similarity 3, member C |
| chr5_-_64818813 | 0.03 |
ENSRNOT00000009111
ENSRNOT00000086505 |
Aldob
|
aldolase, fructose-bisphosphate B |
| chr12_-_9864791 | 0.03 |
ENSRNOT00000073026
|
Gtf3a
|
general transcription factor III A |
| chr16_+_51748970 | 0.03 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr17_+_10559680 | 0.03 |
ENSRNOT00000023320
|
Nop16
|
NOP16 nucleolar protein |
| chr3_+_5709236 | 0.03 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr4_-_157486844 | 0.03 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr4_+_148782479 | 0.03 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr17_+_72218769 | 0.03 |
ENSRNOT00000041346
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr5_+_116973159 | 0.03 |
ENSRNOT00000046475
|
LOC100912024
|
uncharacterized LOC100912024 |
| chr2_+_204512302 | 0.03 |
ENSRNOT00000021846
|
Casq2
|
calsequestrin 2 |
| chr1_+_122981755 | 0.03 |
ENSRNOT00000013468
|
Ndn
|
necdin, MAGE family member |
| chr5_-_17061361 | 0.03 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr10_-_4257868 | 0.03 |
ENSRNOT00000035886
|
Tnfrsf17
|
TNF receptor superfamily member 17 |
| chr15_+_61673560 | 0.03 |
ENSRNOT00000093661
|
Mtrf1
|
mitochondrial translation release factor 1 |
| chr18_+_55391388 | 0.02 |
ENSRNOT00000071612
|
LOC100910934
|
interferon-inducible GTPase 1-like |
| chr3_-_36660758 | 0.02 |
ENSRNOT00000006111
|
Rnd3
|
Rho family GTPase 3 |
| chr18_-_67224566 | 0.02 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr5_+_147476221 | 0.02 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr13_-_72063347 | 0.02 |
ENSRNOT00000090544
ENSRNOT00000003869 |
Cacna1e
|
calcium voltage-gated channel subunit alpha1 E |
| chr3_+_150323116 | 0.02 |
ENSRNOT00000023429
ENSRNOT00000080485 |
Raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr13_+_90692666 | 0.02 |
ENSRNOT00000010028
|
Igsf8
|
immunoglobulin superfamily, member 8 |
| chr14_-_21748356 | 0.02 |
ENSRNOT00000002670
|
Cabs1
|
calcium binding protein, spermatid associated 1 |
| chr1_-_199270627 | 0.02 |
ENSRNOT00000026063
|
Stx1b
|
syntaxin 1B |
| chr15_+_17834635 | 0.02 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr12_+_41600005 | 0.02 |
ENSRNOT00000001872
|
Plbd2
|
phospholipase B domain containing 2 |
| chr10_-_56591364 | 0.02 |
ENSRNOT00000032481
|
Elp5
|
elongator acetyltransferase complex subunit 5 |
| chr4_-_164049599 | 0.02 |
ENSRNOT00000078267
|
AABR07062183.1
|
|
| chr2_+_188784222 | 0.02 |
ENSRNOT00000028095
|
Pmvk
|
phosphomevalonate kinase |
| chr1_+_101324652 | 0.02 |
ENSRNOT00000028116
|
Hrc
|
histidine rich calcium binding protein |
| chr4_+_88271061 | 0.02 |
ENSRNOT00000087374
|
Vom1r86
|
vomeronasal 1 receptor 86 |
| chr2_+_256964860 | 0.02 |
ENSRNOT00000073547
|
Ifi44l
|
interferon-induced protein 44-like |
| chr6_+_107460668 | 0.02 |
ENSRNOT00000013515
|
Acot2
|
acyl-CoA thioesterase 2 |
| chr18_+_61563053 | 0.02 |
ENSRNOT00000022845
|
Grp
|
gastrin releasing peptide |
| chr9_-_50820290 | 0.02 |
ENSRNOT00000050748
|
LOC102548013
|
uncharacterized LOC102548013 |
| chr15_+_19603288 | 0.02 |
ENSRNOT00000035491
|
LOC102552640
|
REST corepressor 2-like |
| chr6_-_108120579 | 0.02 |
ENSRNOT00000041163
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr16_+_49266903 | 0.02 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr2_-_261337163 | 0.02 |
ENSRNOT00000030341
|
Tnni3k
|
TNNI3 interacting kinase |
| chrX_+_33895769 | 0.02 |
ENSRNOT00000085993
ENSRNOT00000079365 |
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr10_+_82032656 | 0.02 |
ENSRNOT00000067751
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr3_+_43271596 | 0.02 |
ENSRNOT00000088589
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr1_-_253186695 | 0.02 |
ENSRNOT00000080928
|
AC096809.1
|
|
| chrX_-_62690806 | 0.02 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr2_-_216443518 | 0.02 |
ENSRNOT00000022496
|
Amy1a
|
amylase, alpha 1A |
| chr19_+_9622611 | 0.02 |
ENSRNOT00000061498
|
Slc38a7
|
solute carrier family 38, member 7 |
| chr13_-_90602365 | 0.02 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr1_-_224921092 | 0.02 |
ENSRNOT00000025196
|
Slc3a2
|
solute carrier family 3 member 2 |
| chr1_+_15180328 | 0.02 |
ENSRNOT00000050656
|
Il20ra
|
interleukin 20 receptor subunit alpha |
| chr16_+_2537248 | 0.02 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr11_-_14304603 | 0.02 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr11_-_61470348 | 0.02 |
ENSRNOT00000083841
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr13_+_67545430 | 0.02 |
ENSRNOT00000003405
|
Pdc
|
phosducin |
| chr6_+_132246602 | 0.02 |
ENSRNOT00000009896
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr2_+_198721724 | 0.02 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr9_-_50762082 | 0.01 |
ENSRNOT00000015492
|
Mettl21c
|
methyltransferase like 21C |
| chr18_-_74688551 | 0.01 |
ENSRNOT00000078016
|
Slc14a2
|
solute carrier family 14 member 2 |
| chr13_+_92264231 | 0.01 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr6_-_136550371 | 0.01 |
ENSRNOT00000065971
|
Rd3l
|
retinal degeneration 3-like |
| chr13_-_77896697 | 0.01 |
ENSRNOT00000003452
|
Tnn
|
tenascin N |
| chr2_+_208541361 | 0.01 |
ENSRNOT00000021288
|
Tmigd3
|
transmembrane and immunoglobulin domain containing 3 |
| chr4_-_81241152 | 0.01 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr5_-_101138427 | 0.01 |
ENSRNOT00000058615
|
Frem1
|
Fras1 related extracellular matrix 1 |
| chr9_+_71247781 | 0.01 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr8_+_110982777 | 0.01 |
ENSRNOT00000010992
|
Ky
|
kyphoscoliosis peptidase |
| chr10_+_77755216 | 0.01 |
ENSRNOT00000087765
|
Mmd
|
monocyte to macrophage differentiation-associated |
| chr2_-_5577369 | 0.01 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr7_-_104486710 | 0.01 |
ENSRNOT00000084764
|
Gsdmc
|
gasdermin C |
| chr15_-_65507968 | 0.01 |
ENSRNOT00000046240
|
Calm2
|
calmodulin 2 |
| chr1_+_84411726 | 0.01 |
ENSRNOT00000025303
|
Akt2
|
AKT serine/threonine kinase 2 |
| chr15_-_37325178 | 0.01 |
ENSRNOT00000011699
|
Gja3
|
gap junction protein, alpha 3 |
| chr8_+_1459526 | 0.01 |
ENSRNOT00000034503
|
Kbtbd3
|
kelch repeat and BTB domain containing 3 |
| chr20_+_20577561 | 0.01 |
ENSRNOT00000081113
|
Cdk1
|
cyclin-dependent kinase 1 |
| chr6_-_136145837 | 0.01 |
ENSRNOT00000015122
|
Ckb
|
creatine kinase B |
| chr15_-_28314459 | 0.01 |
ENSRNOT00000042055
ENSRNOT00000040540 |
Ndrg2
|
NDRG family member 2 |
| chr18_+_35574002 | 0.01 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr1_-_259287684 | 0.01 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr12_-_29952196 | 0.01 |
ENSRNOT00000086175
|
Tmem248
|
transmembrane protein 248 |
| chrX_-_136150209 | 0.01 |
ENSRNOT00000048498
|
Enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr3_+_56056925 | 0.01 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr8_+_22750336 | 0.01 |
ENSRNOT00000013496
|
Ldlr
|
low density lipoprotein receptor |
| chrX_-_62698830 | 0.01 |
ENSRNOT00000076359
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr15_-_45550285 | 0.01 |
ENSRNOT00000012948
|
Gucy1b2
|
guanylate cyclase 1 soluble subunit beta 2 |
| chr4_-_23718047 | 0.01 |
ENSRNOT00000011359
|
RGD1564345
|
similar to RIKEN cDNA 4921511H03 |
| chr5_+_154598758 | 0.01 |
ENSRNOT00000015776
|
Tcea3
|
transcription elongation factor A3 |
| chr3_-_14112851 | 0.01 |
ENSRNOT00000092736
|
C5
|
complement C5 |
| chr13_-_30800451 | 0.01 |
ENSRNOT00000046791
|
Rpl21
|
ribosomal protein L21 |
| chr5_+_135442031 | 0.01 |
ENSRNOT00000090852
ENSRNOT00000034464 |
Ccdc17
|
coiled-coil domain containing 17 |
| chr1_-_56683731 | 0.01 |
ENSRNOT00000014552
|
Thbs2
|
thrombospondin 2 |
| chr2_+_226563050 | 0.01 |
ENSRNOT00000065111
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr17_-_10559627 | 0.01 |
ENSRNOT00000023515
|
Higd2a
|
HIG1 hypoxia inducible domain family, member 2A |
| chr9_+_37727942 | 0.01 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr10_+_89459650 | 0.01 |
ENSRNOT00000028149
ENSRNOT00000077942 |
Nbr1
|
NBR1, autophagy cargo receptor |
| chr6_+_29061618 | 0.01 |
ENSRNOT00000092223
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr5_+_15043955 | 0.01 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chr9_+_118586179 | 0.01 |
ENSRNOT00000022351
|
Dlgap1
|
DLG associated protein 1 |
| chr1_-_64090017 | 0.01 |
ENSRNOT00000086622
ENSRNOT00000091654 |
Rps9
|
ribosomal protein S9 |
| chr8_-_132919110 | 0.01 |
ENSRNOT00000008747
|
Xcr1
|
X-C motif chemokine receptor 1 |
| chr16_-_31301880 | 0.01 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr10_+_34383396 | 0.01 |
ENSRNOT00000047186
|
Olr1386
|
olfactory receptor 1386 |
| chr5_+_33097654 | 0.01 |
ENSRNOT00000008087
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chrX_-_64726210 | 0.01 |
ENSRNOT00000076012
ENSRNOT00000086265 |
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr13_-_84694575 | 0.01 |
ENSRNOT00000066609
|
Fmo13
|
flavin-containing monooxygenase 13 |
| chr3_+_72063504 | 0.01 |
ENSRNOT00000066606
|
Btbd18
|
BTB domain containing 18 |
| chr1_+_218945508 | 0.01 |
ENSRNOT00000034240
|
RGD1311946
|
similar to RIKEN cDNA 1810055G02 |
| chr17_-_27279260 | 0.01 |
ENSRNOT00000018508
|
Snrnp48
|
small nuclear ribonucleoprotein U11/U12 subunit 48 |
| chr10_-_25847994 | 0.01 |
ENSRNOT00000082076
|
Mat2b
|
methionine adenosyltransferase 2B |
| chrX_-_104814145 | 0.01 |
ENSRNOT00000004909
|
Sytl4
|
synaptotagmin-like 4 |
| chr1_-_166912524 | 0.01 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr7_+_40316639 | 0.00 |
ENSRNOT00000080150
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chrX_-_135041027 | 0.00 |
ENSRNOT00000006484
|
Zdhhc9
|
zinc finger, DHHC-type containing 9 |
| chr12_+_30655898 | 0.00 |
ENSRNOT00000079144
|
Zfp11
|
zinc finger protein 11 |
| chr1_-_64350338 | 0.00 |
ENSRNOT00000078444
|
Cacng8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chr2_+_198655437 | 0.00 |
ENSRNOT00000028781
|
Hfe2
|
hemochromatosis type 2 (juvenile) |
| chr17_-_47394231 | 0.00 |
ENSRNOT00000079368
ENSRNOT00000079216 |
Sfrp4
|
secreted frizzled-related protein 4 |
| chr14_-_72025137 | 0.00 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr13_+_93751759 | 0.00 |
ENSRNOT00000065706
|
Wdr64
|
WD repeat domain 64 |
| chr1_-_217902473 | 0.00 |
ENSRNOT00000028329
|
Ano1
|
anoctamin 1 |
| chr7_-_107391184 | 0.00 |
ENSRNOT00000056793
|
Tmem71
|
transmembrane protein 71 |
| chr1_+_167944448 | 0.00 |
ENSRNOT00000087667
|
Olr56
|
olfactory receptor 56 |
| chr10_-_71439540 | 0.00 |
ENSRNOT00000087769
|
Tada2a
|
transcriptional adaptor 2A |
| chr8_+_117062884 | 0.00 |
ENSRNOT00000082452
ENSRNOT00000071540 |
Nicn1
|
nicolin 1 |
| chr4_-_62840357 | 0.00 |
ENSRNOT00000059892
|
Slc13a4
|
solute carrier family 13 member 4 |
| chr10_-_15459897 | 0.00 |
ENSRNOT00000027518
|
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr13_-_80819218 | 0.00 |
ENSRNOT00000072922
|
Fmo6
|
flavin containing monooxygenase 6 |
| chr17_+_18031228 | 0.00 |
ENSRNOT00000081708
|
Tpmt
|
thiopurine S-methyltransferase |
| chr2_+_119197239 | 0.00 |
ENSRNOT00000048030
|
Usp13
|
ubiquitin specific peptidase 13 |
| chr10_-_51576376 | 0.00 |
ENSRNOT00000004829
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr15_+_42653148 | 0.00 |
ENSRNOT00000022095
|
Clu
|
clusterin |
| chr9_+_88110731 | 0.00 |
ENSRNOT00000088677
|
Rhbdd1
|
rhomboid domain containing 1 |
| chr9_-_63641400 | 0.00 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr3_-_173944396 | 0.00 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr1_+_78800754 | 0.00 |
ENSRNOT00000084601
|
Dact3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr15_+_39779648 | 0.00 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr18_+_28594892 | 0.00 |
ENSRNOT00000067702
|
Ube2d2
|
ubiquitin-conjugating enzyme E2D 2 |
| chr18_-_31082101 | 0.00 |
ENSRNOT00000084735
ENSRNOT00000060417 |
Hdac3
|
histone deacetylase 3 |
| chr5_+_58995249 | 0.00 |
ENSRNOT00000023411
|
Ccdc107
|
coiled-coil domain containing 107 |
| chr15_+_50891127 | 0.00 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chrX_+_65566047 | 0.00 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0044393 | TCR signalosome(GO:0036398) microspike(GO:0044393) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |